Flu Virus
Creative Biolabs was founded by a group of skilled scientists who have specialized in antibody development for decades. After a large number of successful projects, we are proud of ourselves in being able to provide high quality antibodies with affordable prices. With the extensive experience in hybridoma, CHO, and HEK-293 cells, our custom antibody production service does not only offer you antibodies, but provides the complete support for all steps in antibody production.
Introduction of Flu Virus
Influenza viruses (Flu virus) belong to the family Orthomyxoviridae that is a family of RNA viruses. The genome of the family contains segmented negative-sense single-strand RNA segments. There are seven genera of this family include Influenza A virus, Influenza B virus, Influenza C virus, Influenza D virus, Isavirus, Thogotovirus, and Quaranjavirus. However, only the first four genera can cause influenza in vertebrates including birds, humans, and other mammals. Isaviruses infect salmon. The thogotoviruses are arboviruses to infect vertebrates and invertebrates, such as ticks and mosquitoes. The genome segments of influenza A and B viruses are loosely encapsidated by the nucleoprotein. The polymerase complexes are composed of three polymerase proteins PB1, PB2, and PA located at the ends of the nucleocapsids.
These helical capsids are surrounded by M1 matrix protein and lipid bilayers from the host, in which the virus surface glycoproteins haemagglutinin (HA) and neuraminidase (NA), and the M2 matrix protein are embedded.
Influenza virus as an enveloped virus presents relatively vulnerable to damaging environmental factors. However, it can maintain alive to several hours to several months in water at low temperatures (e.g. <20°C) according to different environmental conditions (such as the humidity and temperature). Flu virus is also sensitive to lipid solvents and detergents, to heat and a low pH. The most typical feature of influenza viruses is their rapid evolution that can result in its great variability. Based on the antigenic properties of envelope proteins, influenza A viruses are subdivided into various subtypes. To date, there are 16 different HA and 9 different NA subtypes. Influenza B viruses have not been divided into subtypes. The accumulation of point mutations generally causes a step-by-step modification of the virus proteins. However, the variability of the type B viruses is also characterized by other mechanisms such as insertion and deletion, as the influenza B lines can remain co-circulating and stable for more than 20 years. Influenza A/H5N1 subtype A is a severe flu virus infection that has caused the deaths in humans that led to special attention to the public. This is a new influenza A virus that can cause deadly infections in humans. Human H5N1 transmissions have caused high mortality in 10 countries since it first outbroke in poultry in South-East Asia in 2003. The virus has the potential to better adapt to humans and then be able to spread in the human population.
 Fig.1 Influenza virus.
Fig.1 Influenza virus.
Taxonomy
There are four genera of influenza virus, each containing only a single species, or type. Influenza A and C infect a variety of species, while influenza B almost exclusively infects humans, and influenza D infects cattle and pigs.
- Influenza A
Influenza A viruses are further divided into Sixteen H subtypes (or serotypes) and nine N subtypes based on the viral surface proteins hemagglutinin (HA or H) and neuraminidase (NA or N). Type A viruses represent the most virulent human pathogens among the three influenza types that can cause the most severe disease. The serotypes outbroke and caused high mortality in humans, including H1N1 caused "Spanish flu" in 1918 and "Swine flu" in 2009, H2N2 caused "Asian Flu", H3N2 caused "Hong Kong Flu", H5N1 caused "avian" or "bird flu", H7N7 caused zoonotic potential, H1N2 caused endemic in humans and pigs, etc.
- Influenza B
Influenza B virus is exclusively a human pathogen and mutates at a rate 2-3 times lower than type A. Thus, it is less genetically diverse with only one influenza B serotype. Due to the lack of antigenic diversity, immunity to influenza B is usually acquired at a very young age. This reduced rate of antigenic change and limited host range ensures that there is no influenza B pandemic.
- Influenza C
The influenza C virus can infect both humans and pigs, and cause severe illness and local epidemics. However, type C influenza is less common than other types of influenza and usually causes mild illness in children.
- Influenza D
Influenza D was classified in 2016 and first isolated in 2011. This genus appears to be most closely related to Influenza C. There are at least two strains of this genus and can infect cattle and pigs.
Morphology and Structure
The virion is pleomorphic and the envelope is spherical or filamentous form. The diameter of the virus particles is approximately 80 to 120 nm. The major glycoprotein (HA) is embedded irregularly by clusters of neuraminidase (NA), with a ratio of about 4-5 to 1 of HA /NA. The nucleocapsids are coated by cholesterol-laden membranes with protruding glycoproteins. Nucleoproteins present different size classes with a loop at each end. The ribonucleoproteins are filamentous, 50 to 130 nm long and 9 to 15 nm in diameter.
The influenza A virus usually forms both ellipsoidal, bacilliform, and filamentous particles. The genome of influenza A contains eight pieces of segmented negative-sense RNA (13.5 kilobases), which can encode 11 proteins (HA, NA, NP, M1, M2, NS1, NEP, PA, PB1, PB1-F2, PB2). Hemagglutinin and neuraminidase are well-studied viral glycoprotein proteins located on the outside of the viral particles. Neuraminidase as an enzyme functions in the release of progeny virus from infected cells by cleaving sugars from viral particles. Hemagglutinin is a lectin that plays a role in regulating the binding of the virus to target cells and entry of the viral genome into the target cell. So far, the hemagglutinin (H) and neuraminidase (N) proteins have been regarded as targets for the development of antiviral drugs, and antibodies production that are used to classify the different serotypes of influenza A viruses.
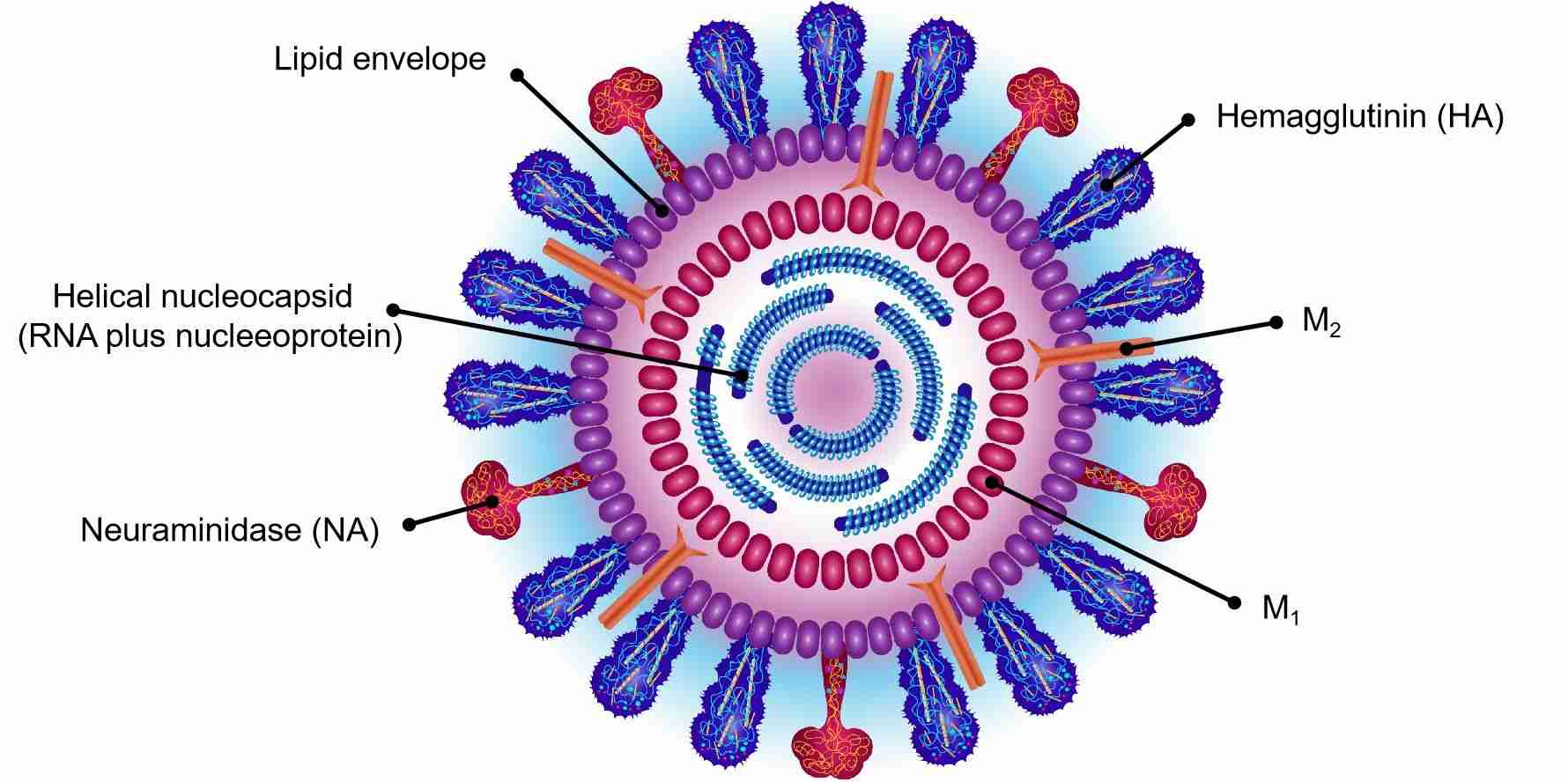 Fig.2 Structure of influenza virus.
Fig.2 Structure of influenza virus.
Epidemiology
Since 1977, the subtypes influenza A/H3N2, A/H1N1 and influenza B have been found with a co-circulation in humans. The infection of subtype H1N2 was first observed in humans in 1977, and then it caused local outbreaks in some countries. Generally, human influenza virus infections present a world-wide distribution. Seasonal influenza epidemics regularly outbreaks in the Northern and the Southern hemispheres each winter, which causes approximately 500,000 deaths per year world-wide. Approximately 10-15% of the population is affected during the annual influenza epidemics. The influenza epidemics also vary according to the degree of severity. In the past years, the seasons of 2002/2003 and 2004/2005 were characterized by extraordinarily high influenza virus activity, which have caused approximately 2,900,000 excess consultations, 900,000 sick, 14,000 additional hospitalizations, and approximately 10,000 deaths. Most influenza related deaths are associated with age groups over 60.
Influenza pandemics are characterized by the occurrence (recurrence) of influenza A subtype due to most of the human population is not immune to it and can cause a worldwide epidemic. There are three main pandemics over the past century, including the Spanish Flu of 1918 (H1N1) caused about 40 million deaths, the Asian influenza of 1957 (H2N2) caused 1-2 million death and the Hong Kong Flu of 1968 (H2N3) caused 0.75-1 million deaths. It is suspected that the next pandemic may also be caused by the H5N1 subtype that is highly pathogenic influenza virus. Since the first human H5N1 outbreaks in 2003, it has caused 331 infections and 202 deaths. At present, Indonesia and Egypt have become two endemic areas with recurrent human infections and deaths.
Replication Cycle
Influenza is usually transmitted by infected mammals through the air through coughing or sneezing, producing aerosols containing the virus, and by infected birds through feces. Influenza can also be transmitted through saliva, nasal secretions, feces and blood.
Out of a host, the influenza virus can remain infectious for about a week under human body temperature, for more than 30 days under 0°C (32°F), and for an indefinite period under extremely low temperature. However, they also can be inactivated easily by disinfectants and detergents.
The virus binds to a cell through the interaction of hemagglutinin and sialic acid sugars that locate on the surfaces of epithelial cells of the lung and throat. Then, the virus is imported into the cell by endocytosis. Upon enters the acidic endosome, part of the haemagglutinin protein fuses the viral envelope with the vacuole's membrane. Subsequently, the release of the viral RNA (vRNA) molecules, accessory proteins and RNA-dependent RNA polymerase into the cytoplasm can form a complex that is transported into the cell nucleus, where the RNA-dependent RNA polymerase begins transcribing complementary positive-sense cRNA. The cRNA is exported into the cytoplasm, translated or remains in the nucleus. The newly synthesized viral proteins are either secreted to the cell surface through Golgi apparatus, or transported back to the nucleus to combine with vRNA to form new viral genome particles. Other viral proteins are committed to degrading cellular mRNA and using the released nucleotides for vRNA synthesis and also inhibiting the translation of host-cell mRNAs.
Negative-sense vRNAs become the genomes of future viruses, and RNA-dependent RNA transcriptase, and other viral proteins are assembled into a virion. Hemagglutinin and neuraminidase molecules form a bulge in the cell membrane. The vRNA and viral core proteins leave the nucleus and enter this membrane protrusion. The mature virus germinates from the cell in the sphere of the host phospholipid membrane, with which hemagglutinin and neuraminidase are obtained. Upon the release of a new influenza virus, the host cell dies.
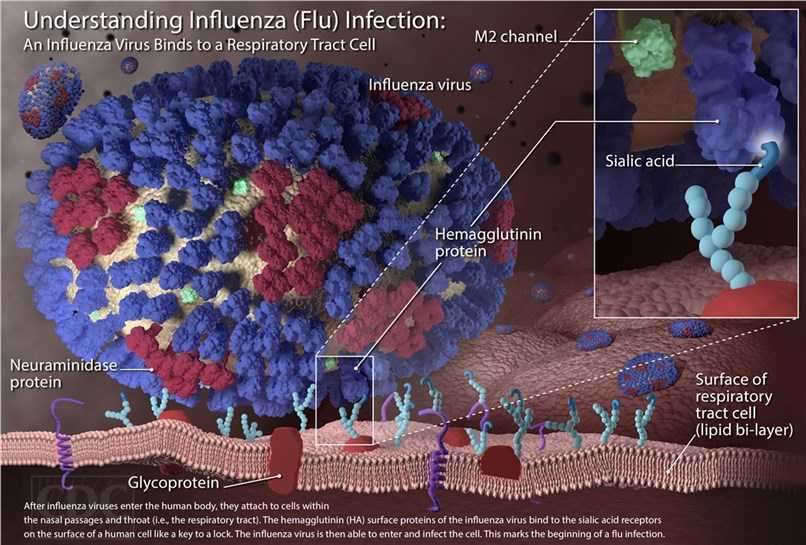 Fig.3 The mechanism of influenza virus infection.
Fig.3 The mechanism of influenza virus infection.
Detection Methods
A number of methods can be used for the detection of an influenza virus infection.
- Serology
Specific antibodies against influenza virus in the serum can be detected as total antibody levels or differentiated as single antibody sub-classes (IgA, IgM, IgG). Traditional methods include complement-binding reaction (CBR), enzyme immunoassay (EIA) and immunofluorescence test (IFT). Some more sophisticated methods such as the haemagglutination inhibition test (HIT) and the neutralization test commonly require specialized reagents and are performed by high-standard laboratories.
- Virus Culture
Isolation of influenza virus required specialized technology, which is only performed by laboratories under biosafety level 2 conditions. Virus culture is very time-consuming and requires the presence of replicable virus particles in patient samples. Embryonated hen eggs as common material have been used for culturing for decades. Madin-Darby canine kidney (MDCK) cells have found to be particularly suitable. The occurrence of a cytopathic effect may take days to two weeks. Following this, the culture can be confirmed by a specific test (such as ELISA or IFT).
- Antigen Detection
Antigen detection tests can be performed faster in any laboratory compared with virus culturing. For this detection, IFT or ELISA tests can be easily performed and can provide results in only a few hours. These methods show relatively good results if there is a sufficient amount of virus antigen available in the patient’s sample.
- Genome Detection
For genome detection, the Polymerase chain reaction (PCR) is a very sensitive and specific method. With the aid of specific oligonucleotide primers, the required genome sequences are amplified that they can subsequently be detected using various techniques. Higher specificity of PCR can be obtained by using specific probes by means of hybridization blot or PCR-EIA. Using nested PCR can achieve higher specificity.
Therapy and Prophylaxis
For the prophylaxis and treatment of influenza virus infections, vaccines and drugs are useful. Vaccines consist of either inactivated or live-attenuated virions of the H1N1 and H3N2 human influenza A viruses, as well as influenza B viruses. Due to the evolution of the antigenicities of the wild viruses, vaccines are also reformulated annually by updating the seed strains. Vaccines will lose the ability to protect the vaccinees upon the antigenicities of the seed strains and wild viruses do not match. Besides, escape mutants often occur.
Drugs used for the treatment of influenza include Amantadine and Rimantadine, which can inhibit the uncoating of virions by interfering with M2, and Oseltamivir (Tamiflu), Zanamivir, and Peramivir, which inhibit the release of virions from infected cells by interfering with NA. However, escape mutants are also often generated.
Fig.4 Oseltamivir prophylaxis reduces inflammation-induced morbidity while maintaining establishment of functional cross-strain protective CD8+ T cell memory pools.1
Anti-influenza virus antibodies are useful for diagnosis and investigations of virus. Creative Biolabs provides a full range of antibodies against the Influenza A Virus Neuraminidase N2, Influenza A Virus Nucleoprotein, Influenza A Virus Neuraminidase N1, Influenza A Virus Hemagglutinin H3, etc. Creative Biolabs has been specialized in developing and producing antibodies for more than ten years, so we are confident that we have expertise, experience as well as enabling technologies on antigen design and antibody purification to customize the high-quality antibodies that you are needed. If you are interested in our antibodies and services, please contact us.
Reference
-
Bird, Nicola L., et al. "Oseltamivir prophylaxis reduces inflammation and facilitates establishment of cross-strain protective T cell memory to influenza viruses." PLoS One 10.6 (2015): e0129768.
Distributed under Open Access license CC BY 4.0, without modification.


