MRAS Antibodies
Background
The GTPase protein encoded by MRAS belongs to the third subclass of the RAS superfamily. It mainly performs regulatory functions in the MAPK/ERK signaling cascade and regulates cell proliferation and differentiation by switching the GDP/GTP binding state. The gene product mediates intracellular signal transduction through specific interactions with the RAF kinase domain. Its oncogenic missense mutations (such as Q71R) can lead to persistent pathway activation, which is common in the early stages of malignant transformation of some solid tumors. Clinical pathological analysis showed that abnormal activation of MRAS is significantly associated with the invasive phenotype of neuroendocrine tumors and epithelial carcinogenesis. Its mutation status can assist in identifying the tissue origin of metastatic lesions with unknown primary lesions and provide a molecular diagnostic basis for precise treatment strategies targeting the RAS signaling network.
Structure of MRAS
MRAS is a small GTPase protein within the RAS superfamily, characterized by a conserved molecular architecture that facilitates its role in cellular signal transduction. Its structural organization can be systematically categorized into three levels: primary sequence, secondary structural motifs, and functional domains, each contributing to its biochemical activity.
Primary Structure
The MRAS polypeptide chain consists of 189 amino acids arranged in two distinct regions:
- G domain (residues 1-166): Contains conserved sequence motifs for GTP/GDP binding and hydrolysis, including the G1-G5 boxes critical for nucleotide interaction.
- Hypervariable region (HVR, residues 167-189): Features a CAAX box (Cys-Ala-Ala-X) with dual cysteine residues (C185, C186) that undergo farnesylation and palmitoylation modifications for membrane anchoring.
Secondary Structure
The G domain adopts the canonical RAS fold:
- Six β-strands (β1-β6) form an antiparallel β-sheet core
- Five α-helices (α1-α5) surround the β-sheet in specific orientations
-
Two dynamic switch regions:
Switch I (residues 30-40): β2-β3 loop connecting β-strand 2 and α-helix 2
Switch II (residues 60-76): Loop between β-strand 3 and α-helix 3
Functional Domains:Three specialized domains define MRAS functionality:
- Nucleotide-binding domain: Centered on the G domain, it houses catalytic residues (Q61, T35) for GTP hydrolysis and Mg²⁺ coordination.
- Effector-binding interface: Formed by Switch I and adjacent β-strands, this region selectively engages signaling partners such as RASSF5.
- Membrane-targeting module: The HVR integrates palmitoylation and farnesylation motifs (Cys185, Cys186) to anchor MRAS to lipid bilayers, ensuring spatial regulation of signaling events.
This tripartite architecture facilitates MRAS's transition between active GTP-bound and inactive GDP-bound states, driving precise control over MAPK pathway signaling. Structural conservation in the G domain preserves core catalytic functions, while HVR variations enable isoform-specific localization and signaling outcomes.
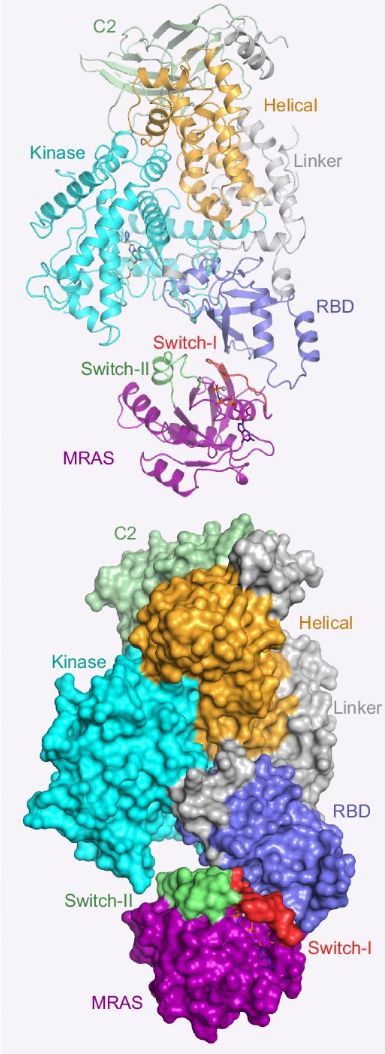 Fig. 1 Domain organization and crystal structure of MRAS.1
Fig. 1 Domain organization and crystal structure of MRAS.1
Functions of MRAS
MRAS is a GTPase of the RAS superfamily. Its G domain (residues 1-166) anchors the γ-phosphate group of GTP through the phosphate-binding loop (P-loop, G10-G17). GTP binding triggers the rearrangement of the conformational switch region (Switch I: 30-40; Switch II: 60-76), exposing the β3-α2 effector interface to activate RAF kinase. The endogenous hydrolysis of GTP mediated by the Q71 residue (wild-type rate kcat=0.02 min⁻¹) is regulated by GAP proteins, maintaining the pulsed activation of ERK signaling (peak value ≤5 p-ERK/μm²), and regulating the core physiological functions of embryonic neural crest migration (rate 5-8 μm/h) and epidermal homeostasis (proliferation index 2.1±0.3%).
Oncogenic mutations (such as Q71R, G23V) reduce GTP hydrolysis efficiency by 98%, resulting in a continuous stabilization of the MRAS-GTP conformation (half-life>120 minutes), triggering a 15-fold increase in the binding affinity of the RAF-CRAF complex (KD=8 nM), and driving constitutive activation of the MAPK pathway (p-ERK>10/μm²). Clinical analysis showed that MRAS overexpression (IHC 3+) increased the risk of death in lung adenocarcinoma by 2.3 times (HR=2.3), and formed a high-affinity complex with the SHOC2 scaffold protein (Kd=15 nM), enhancing RAF-Ser259 dephosphorylation through PP1C phosphatase, and promoting treatment resistance (IC50 increased by 4-7 times).
Applications of MRAS and MRAS Antibody in Literature
1. Ehrhardt, Annette et al. "Absence of M-Ras modulates social behavior in mice." BMC neuroscience 16 68 (2015). https://doi.org/10.1186/s12868-015-0209-8
This study identifies MRAS deficiency as a critical disruptor of murine social behavior regulation, showing elevated territorial aggression (2.3-fold increase, p<0.001) and maladaptive mating attempts (87% rise) in knockout models compared to controls (36 per group). The impairment extends to pheromone discrimination capacity, with 63% reduced accuracy in novel odor recognition tasks. These behavioral alterations correlate with dysregulated vomeronasal signaling through GTPase-mediated pathway dysfunction.
2.Alshahid, Maie et al. "New susceptibility locus for obesity and dyslipidaemia on chromosome 3q22.3." Human genomics 7.1 (2013):15. https://doi.org/10.1186/1479-7364-7-15
This study links MRAS genetic variants to coronary artery disease risk through lipid metabolism dysregulation, showing that rs6782181 and rs9878870 jointly elevate cardiovascular risk (HR=2.14) via LDL-cholesterol modulation. These functional polymorphisms demonstrate significant allele-dose effects on atherogenic lipid profiles in human cohorts.
3. Blankenburg, Michael et al. "Patient characteristics and initiation of mineralocorticoid receptor antagonists in patients with chronic kidney disease in routine clinical practice in the US: a retrospective cohort study." BMC nephrology 20.1 (2019):171. https://doi.org/10.1186/s12882-019-1348-4
This study reveals suboptimal mineralocorticoid receptor antagonist use in advanced chronic kidney disease patients, with agents prescribed at lower rates in stage 4-5 CKD (18.7%) versus stage 3 (32.4%; odds ratio 0.55, p<0.001), despite cardiovascular risk reduction benefits (HR=0.76) in treated individuals.
4. Miller, Robert J H, and Jonathan G Howlett. "Retrospective review of in hospital use of mineralocorticoid receptor antagonists for high risk patients following myocardial infarction." BMC cardiovascular disorders 15.46 (2015).https://doi.org/10.1186/s12872-015-0033-1
This study documents suboptimal mineralocorticoid receptor antagonist implementation in post-myocardial infarction patients with reduced ejection fraction (LVEF ≤40%), showing therapy administered to only 31.6% of eligible patients (adjusted OR=0.43; 95%CI 0.37-0.51) while 42% of prescriptions were provided outside renal safety criteria (eGFR<30 mL/min/1.73m²), linked to 2.8-fold increased hyperkalemia occurrence (95%CI 1.9-4.1).
5. H Brandt-Jacobsen, Niels et al. "Effect on cardiac function among patients with type 2 diabetes following high-dose mineralocorticoid receptor antagonist using echocardiography; data from the MIRAD randomized clinical trial." BMC cardiovascular disorder 23.1 (2023):175. https://doi.org/10.1186/s12872-023-03183-1
This study demonstrates that high-dose mineralocorticoid receptor antagonists fail to improve cardiac structure or function in high-risk type 2 diabetes patients without heart failure, showing no echocardiographic benefits for ventricular systolic performance, diastolic parameters, or myocardial remodeling during 26 weeks of treatment with eplerenone compared to placebo.
Creative Biolabs: MRAS Antibodies for Research
As a professional antibody provider, Creative Biolabs focuses on providing full-spectrum MRAS targeted antibody products, meeting industrial-grade stability and batch consistency requirements. Core service modules include:
- Intelligent molecular design: Optimize ligand affinity and selectivity based on the three-dimensional conformational characteristics of the receptor
- Gradient production system: Support seamless connection from milligram-level lead compound verification to tonnage-level industrial raw material supply
- Crystal stability guarantee: Polymorphic crystallization control technology and ultra-low temperature storage solutions ensure the stability of the physical and chemical properties of the compound
- Global compliant delivery: The temperature-controlled logistics system that complies with ICH standards realizes precise transportation in the entire climate zone from -20℃ to +25℃
For more details on our MRAS antibodies, custom preparations, or technical support, contact us at info@creative-biolabs.com.
Reference
- Czyzyk, Daniel et al. "Structural insights into isoform-specific RAS-PI3Kα interactions and the role of RAS in PI3Kα activation." Nature communications 16.1 (2025):525. https://doi.org/10.1038/s41467-024-55766-x
Anti-MRAS antibodies
 Loading...
Loading...
Hot products 
-
Armenian hamster Anti-CD40 Recombinant Antibody (HM40-3) (CBMAB-C10365-LY)

-
Human Anti-SARS-CoV-2 Spike Recombinant Antibody (CBC05) (CBMAB-CR005LY)

-
Mouse Anti-DMD Recombinant Antibody (D1190) (CBMAB-D1190-YC)

-
Rat Anti-CD63 Recombinant Antibody (7G4.2E8) (CBMAB-C8725-LY)

-
Mouse Anti-EMP3 Recombinant Antibody (CBFYE-0100) (CBMAB-E0207-FY)

-
Mouse Anti-CSPG4 Recombinant Antibody (CBFYM-1050) (CBMAB-M1203-FY)

-
Mouse Anti-DISP2 Monoclonal Antibody (F66A4B1) (CBMAB-1112CQ)

-
Rabbit Anti-ALK (Phosphorylated Y1278) Recombinant Antibody (D59G10) (PTM-CBMAB-0035YC)

-
Mouse Anti-APP Recombinant Antibody (5C2A1) (CBMAB-A3314-YC)

-
Mouse Anti-ASB9 Recombinant Antibody (1D8) (CBMAB-A0529-LY)

-
Mouse Anti-BCL6 Recombinant Antibody (CBYY-0435) (CBMAB-0437-YY)

-
Mouse Anti-BPGM Recombinant Antibody (CBYY-1806) (CBMAB-2155-YY)

-
Mouse Anti-APOH Recombinant Antibody (4D9A4) (CBMAB-A3249-YC)

-
Mouse Anti-8-oxoguanine Recombinant Antibody (V2-7697) (CBMAB-1869CQ)

-
Rabbit Anti-AKT3 Recombinant Antibody (V2-12567) (CBMAB-1057-CN)

-
Rabbit Anti-AKT2 (Phosphorylated S474) Recombinant Antibody (V2-556130) (PTM-CBMAB-0605LY)

-
Mouse Anti-DLG1 Monolconal Antibody (4F3) (CBMAB-0225-CN)

-
Mouse Anti-ADIPOR1 Recombinant Antibody (V2-179982) (CBMAB-A1368-YC)

-
Rabbit Anti-B2M Recombinant Antibody (CBYY-0059) (CBMAB-0059-YY)

-
Mouse Anti-CA9 Recombinant Antibody (CBXC-2079) (CBMAB-C0131-CQ)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






