PPAT Antibodies
Background
PPAT is a key metabolic enzyme widely present in eukaryotes and bacteria. As A core component of the coenzyme A biosynthesis pathway, this enzyme directly regulates important physiological processes such as fatty acid synthesis and cellular signal transduction by catalyzing the phosphoacyl thiol transfer reaction. In 2001, researchers successfully resolved the three-dimensional structure of PPAT through X-ray crystallography. This breakthrough not only clarified its catalytic mechanism but also provided precise molecular targets for the development of new antibiotics, which is of great significance for addressing the increasingly severe problem of drug-resistant bacterial infections. At present, PPAT has become an important model system for studying metabolic regulation and developing antibacterial drugs.
Structure of PPAT
PPAT is a key metabolic enzyme with a molecular weight of approximately 50 kDa, which varies slightly among different species. It plays a core role in the de novo purine biosynthesis pathway of purine nucleotides, catalyzing the first rate-limiting step of this pathway, converting 5-phosphoribose pyrophosphate (PRPP) to 5-phosphoribose amine (PRA), while using glutamine to provide the amino group.
| Species | Human | Mouse | Fruit fly | Yeast | Bacteria |
| Molecular Weight (kDa) | 50 | 49 | 48 | 52 | 47 |
| Primary Structural Differences | Highly expressed in proliferating tissues | Knockout leads to embryo death | Purine demand is low | Participate in the stress response | Simplify the synthetic pathway |
Due to the core role of PPAT in nucleotide metabolism, it is regarded as a potential target for anti-tumor and anti-proliferative drugs. Inhibiting PPAT can block purine synthesis, thereby suppressing the growth of rapidly dividing cancer cells. Furthermore, the PPAT of certain microorganisms (such as Plasmodium and bacteria) differs from the human version, making it a potential target for the development of anti-infective drugs.
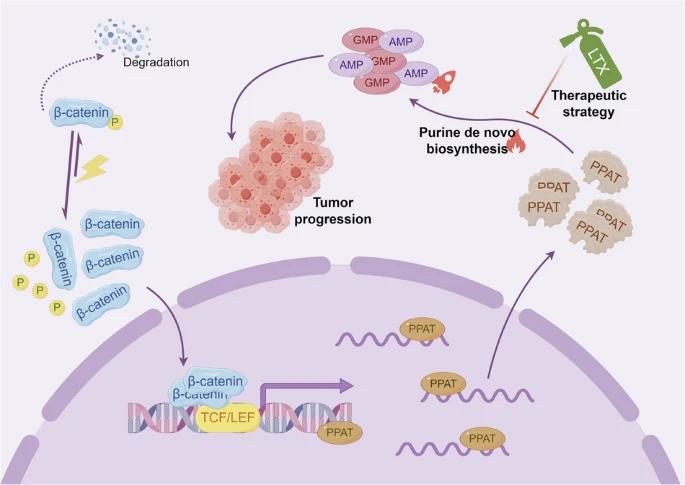 Fig. 1 Targeting PPAT-Mediated Purine Synthesis as a Therapeutic Strategy for β-Catenin-Driven Hepatoblastoma.1
Fig. 1 Targeting PPAT-Mediated Purine Synthesis as a Therapeutic Strategy for β-Catenin-Driven Hepatoblastoma.1
Key structural properties of PPAT:
- Multi-domain α/β folding structure
- Conserved hydrophobic active center
- Key catalytic residues regulate reactions
- Allosteric regulatory sites control enzyme activity
Functions of PPAT
PPAT is the rate-limiting enzyme in the de novo synthesis of purine nucleotides (DNPS) pathway. Its main function is to catalyze the conversion of PRPP (5-phosphoribose pyrophosphate) to PRA (5-phosphoribose amine), thereby regulating the supply of purine nucleotides within cells. In addition, PPAT also plays an important role in processes such as cell proliferation, tumor metabolic reprogramming, and microbial infection.
| Function | Description |
| Purine nucleotide synthesis | Catalyze the first rate-limiting reaction of DNPS, affecting the generation of AMP/GMP and maintaining the balance of the cellular nucleotide pool. |
| Regulation of cell proliferation | In rapidly dividing cells, such as cancer cells, immune cells) increased, provide the necessary DNA/RNA synthesis of purines precursor. |
| Tumor metabolic dependence | Many cancers (such as hepatoblastoma) rely on the purine synthesis pathway mediated by PPAT, making it a potential therapeutic target. |
| Microorganisms must survive | Bacterial and parasitic PPAT differ from the human version and can be used as specific targets for anti-infective drugs. |
| Allosteric adjustment | Inhibited by AMP/GMP feedback, it prevents excessive accumulation of purine and maintains metabolic homeostasis. |
The catalytic efficiency of PPAT is regulated by both the substrate PRPP concentration and the inhibition of the final product. Its enzyme kinetics curve shows typical allosteric regulation characteristics (non-Michaelis kinetics), ensuring the precise matching of purine synthesis with cellular requirements. Under pathological conditions (such as β-catenin mutant tumors), the expression of PPAT abnormally increases, resulting in excessive synthesis of purine nucleotides and driving the malignant progression of tumors.
Applications of PPAT and PPAT Antibody in Literature
1. Ding, Ming, et al. "Therapeutic targeting de novo purine biosynthesis driven by β-catenin-dependent PPAT upregulation in hepatoblastoma." Cell Death & Disease 16.1 (2025): 179. https://doi.org/10.1038/s41419-025-07502-6
The article's research found that PPAT was significantly highly expressed in hepatoblastoma (HB), promoting tumor proliferation and metastasis. The mutation β-catenin can activate PPAT transcription, enhance the purine synthesis pathway, and form a metabolic weakness of HB. Experiments have confirmed that the DNPS inhibitor lometesol can effectively inhibit the progression of HB. This study provides a new strategy for the targeted therapy of β-catenin mutant HB.
2. Huo, Fu-Chun, et al. "SHMT2 promotes the tumorigenesis of renal cell carcinoma by regulating the m6A modification of PPAT." Genomics 114.4 (2022): 110424.https://doi.org/10.1016/j.ygeno.2022.110424
The article's research found that SHMT2 promotes the generation of endogenous methyl donor SAM through the serine/glycine-carbon metabolic network, enhances m6A modification, and thereby upregulates PPAT expression in an M6A-IGF2BP2-dependent manner. The SHMT2-PPAT axis promotes the proliferation of renal cancer cells by inducing the G1/S phase transition, revealing the key regulatory roles of one-carbon metabolism and purine synthesis in tumor progression.
3. Kitagawa, Yuki, et al. "Phosphoribosyl pyrophosphate amidotransferase: Novel biomarker and therapeutic target for nasopharyngeal carcinoma." Cancer Science 115.11 (2024): 3587-3595. https://doi.org/10.1111/cas.16314
The article's research found that phosphoribose pyrophosphate amide transferase (PPAT), as the rate-limiting enzyme for de novo purine synthesis, is significantly highly expressed in nasopharyngeal carcinoma (NPC), especially in cases positive for Epstein-Barr virus. PPAT knockdown can inhibit the proliferation and invasion of cancer cells, and its expression is regulated by glutamine. The antagonist 6-diazo-5-oxo-L-norleucine can block this pathway. Clinical analysis shows that high expression of PPAT is significantly associated with disease progression and poor prognosis, suggesting its potential as a therapeutic target and prognostic marker.
4. Goswami, Moloy T., et al. "Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer." Oncotarget 6.27 (2015): 23445. https://doi.org/ 10.18632/oncotarget.4352
The article's research found that the rate-limiting enzyme PPAT for purine synthesis and its homologous gene PAICS were abnormally highly expressed in lung adenocarcinoma and were significantly associated with disease progression and poor prognosis. Genetic manipulation experiments have confirmed that PPAT/PAICS promote the proliferation and invasion of cancer cells by regulating the activity of pyruvate kinase. The study also found that there were amplification/aneuploidy variations in the PAT-PAICS genomic region, and the expressions of both were regulated by the glutamide-DON pathway, revealing the key mechanism and potential therapeutic targets of purine metabolism reprogramming in lung adenocarcinoma.
5. Wu, Zheng, et al. "NUDT5 regulates purine metabolism and thiopurine sensitivity by interacting with PPAT." bioRxiv (2025): 2025-03. https://doi.org/ 10.1101/2025.03.29.646096
The article's research found that NUDT5 regulates the nucleotide metabolism balance by directly binding to purine to synthesize the rate-limiting enzyme PPAT. When the purine rescue pathway is activated, the NUDT5-PPAT interaction promotes the depolymerization of purinosome, thereby inhibiting the de novo synthesis pathway. This regulation is independent of the NUDT5 hydrolase activity. Mutations in its interaction sites can lead to purine metabolism disorders and thiopurine drug resistance, revealing the key role of the PPAT-NUDT5 axis in maintaining nucleotide homeostasis.
Creative Biolabs: PPAT Antibodies for Research
Creative Biolabs specializes in the production of high-quality PPAT antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western Blot, Immunohistochemistry (IHC), Immunoprecipitation (IP) and other diagnostic methodologies.
- Custom Myoglobin Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our PPAT antibodies, custom preparations, or technical support, contact us at info@creative-biolabs.com.
Reference
- Ding, Ming, et al. "Therapeutic targeting de novo purine biosynthesis driven by β-catenin-dependent PPAT upregulation in hepatoblastoma." Cell Death & Disease 16.1 (2025): 179. https://doi.org/10.1038/s41419-025-07502-6
Anti-PPAT antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-AGO2 Recombinant Antibody (V2-634169) (CBMAB-AP203LY)

-
Mouse Anti-CGAS Recombinant Antibody (CBFYM-0995) (CBMAB-M1146-FY)

-
Mouse Anti-FLI1 Recombinant Antibody (CBXF-0733) (CBMAB-F0435-CQ)

-
Mouse Anti-AFDN Recombinant Antibody (V2-58751) (CBMAB-L0408-YJ)

-
Rabbit Anti-AKT3 Recombinant Antibody (V2-12567) (CBMAB-1057-CN)

-
Mouse Anti-BACE1 Recombinant Antibody (61-3E7) (CBMAB-1183-CN)

-
Mouse Anti-ACO2 Recombinant Antibody (V2-179329) (CBMAB-A0627-YC)

-
Rat Anti-AChR Recombinant Antibody (V2-12500) (CBMAB-0990-CN)

-
Mouse Anti-2C TCR Recombinant Antibody (V2-1556) (CBMAB-0951-LY)

-
Mouse Anti-ADV Recombinant Antibody (V2-503423) (CBMAB-V208-1364-FY)

-
Mouse Anti-AAV-5 Recombinant Antibody (V2-503416) (CBMAB-V208-1402-FY)

-
Mouse Anti-ALX1 Recombinant Antibody (96k) (CBMAB-C0616-FY)

-
Mouse Anti-ARHGAP5 Recombinant Antibody (54/P190-B) (CBMAB-P0070-YC)

-
Mouse Anti-DDC Recombinant Antibody (8E8) (CBMAB-0992-YC)

-
Mouse Anti-CAT Recombinant Antibody (724810) (CBMAB-C8431-LY)

-
Mouse Anti-DHFR Recombinant Antibody (D0821) (CBMAB-D0821-YC)

-
Mouse Anti-CTCF Recombinant Antibody (CBFYC-2371) (CBMAB-C2443-FY)

-
Mouse Anti-APP Recombinant Antibody (DE2B4) (CBMAB-1122-CN)

-
Mouse Anti-AAV9 Recombinant Antibody (V2-634029) (CBMAB-AP023LY)

-
Mouse Anti-Acetyl SMC3 (K105/K106) Recombinant Antibody (V2-634053) (CBMAB-AP052LY)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot








