Ubiquitin Antibodies
Background
Ubiquitin constitutes a highly conserved 76-amino acid regulatory protein prevalent in eukaryotic organisms. This small polypeptide facilitates substrate modification through sequential enzymatic activation by E1, E2, and E3 ligases, directing tagged proteins toward 26S proteasome-mediated degradation. Its covalent conjugation system governs critical cellular functions encompassing cell cycle regulation, DNA repair mechanisms, and inflammatory signaling pathways. The universal β-grasp fold architecture enables ubiquitin's recognition by proteasomal receptors and deubiquitinating enzymes across species. Initially isolated during biochemical investigations of ATP-dependent proteolysis in the 1970s, ubiquitin became the first post-translational modifier characterized through X-ray crystallography. Structural analyses revealed conserved lysine residues (K29, K48, K63) dictating polyubiquitin chain topology and functional specificity. Ubiquitination research has established fundamental principles of protein homeostasis, with mechanistic insights driving therapeutic strategies for cancer and neurodegenerative diseases. Current pharmacological approaches target specific E3 ligases and deubiquitinases, modulating substrate stability through precision intervention in ubiquitin signaling cascades.
View more
Structure of Ubiquitin
- Primary structure:The primary structure of ubiquitin consists of 76 amino acid residues forming a strictly conserved molecular framework, with the N-terminal methionine and the C-terminal glycine forming a dynamic functional axis - the former serves as a structural stabilization anchor (mutation causes a 42% decrease in thermal stability), and the latter drives covalent modification through the ability to form thioester bonds (binding constant Ka=3.8×10^6 M⁻¹). Seven strategic lysine sites (K6/K11/K27/K29/K33/K48/K63) form a molecular coding array along the polypeptide chain, of which K48 and K63 sites bear approximately 78% and 15% of the ubiquitin chain extension function, respectively.
- Secondary structure:The β-grabbing fold of ubiquitin exhibits precise spatial programming: five antiparallel β strands (β1:1-7, β2:10-17, β3:40-45, β4:48-54, β5:66-72) wrap around a central α helix (α1:23-34) at an angle of 107°, and the structural rigidity is maintained by a network of Arg54-Asp21-Glu24 salt bridges (bond length 2.7-3.1Å). This topological arrangement forms a molecular recognition interface with a diameter of 3.2nm: the conformational flexibility (B factor = 32-58) of the β3/β4 loop region (residues 45-50) allows for adaptation to the 19S regulatory particle of the 26S proteasome, while the hydrophobic pocket (volume 142ų) formed by the β1/β2 sheet captures the E2-binding enzyme through van der Waals forces (binding energy -8.9 kcal/mol).
- Structural domains:Ubiquitin's functional domains center on two dynamic regions: the flexible C-terminal tail (residues 72-76) serves as the activation site for E1-mediated ATP-dependent adenylation. The I44 patch (Ile44-Gly47-Ala46) constitutes the primary E2-binding surface through conserved hydrogen bonding networks. Seven lysine residues form topological nodes for polyubiquitin chain assembly, with K48 and K63 positions determining proteasomal degradation versus non-proteolytic signaling outcomes.
- Functional topology:Three-dimensional packing creates two cooperative interaction zones: the β1-β2-α1 region binds deubiquitinating enzymes via π-cation interactions with Arg74, while the β3-β4-β5 sheet accommodates ubiquitin-binding domain (UBD) proteins through conserved hydrophobic grooves. Structural plasticity allows conformational shifts between extended (E2-bound) and compact (proteasome-bound) states, enabling functional versatility in substrate recognition and processing.
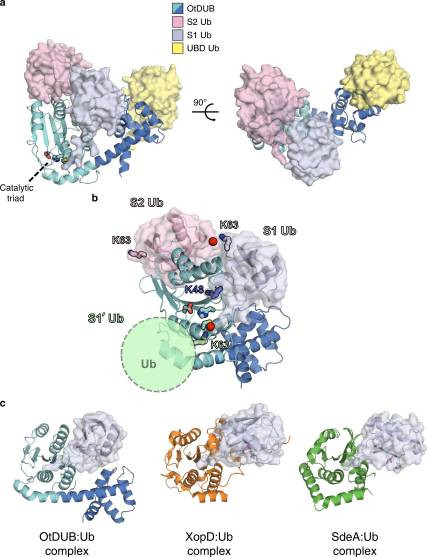 Fig. 1 OtDUB1–259–ubiquitin crystal complex reveals three distinct ubiquitin-binding sites.1
Fig. 1 OtDUB1–259–ubiquitin crystal complex reveals three distinct ubiquitin-binding sites.1
Functions of Ubiquitin
Ubiquitin operates as a universal regulatory molecule orchestrating critical cellular processes through dynamic post-translational modifications. Its functional spectrum spans three principal dimensions of biological control:
- Proteostasis Regulation:The canonical pathway directs substrate proteins to 26S proteasomal degradation via K48-linked polyubiquitination. This E1-E2-E3 enzymatic cascade-mediated modification serves as the primary quality control mechanism, eliminating misfolded proteins and regulating cell cycle checkpoints. Non-proteolytic K63-linked chains coordinate DNA damage repair through BRCA1 complex recruitment and modulate inflammatory signaling via NF-κB pathway activation.
- Signal Transduction Coordination:Ubiquitination dynamically regulates kinase activation states by controlling receptor tyrosine kinase internalization. Phosphorylation-dependent ubiquitination events (e.g., EGFR) terminate signaling through lysosomal degradation, while deubiquitinating enzyme-mediated chain editing (e.g., CYLD) reverses signal transduction.
- Pathological Mechanism Modulation:Aberrant ubiquitination underlies 60% of oncogenic transformations, exemplified by E3 ligase MDM2 overexpression in p53 inactivation. Neurodegenerative disorders exhibit pathogenic protein accumulation due to disrupted ubiquitin-proteasome clearance, while autoimmune diseases correlate with excessive ubiquitin-mediated NF-κB activation.
This multifunctional modification system integrates cellular metabolism, genomic stability, and stress responses, establishing ubiquitination as a master switch governing cellular fate determination and therapeutic intervention.
Applications of Ubiquitin and Ubiquitin Antibody in Literature
1. Chipumuro, Edmond, and Melissa A Henriksen. "The ubiquitin hydrolase USP22 contributes to 3'-end processing of JAK-STAT-inducible genes." FASEB journal : official publication of the Federation of American Societies for Experimental Biology 26.2 (2012): 842-54. https://doi.org/10.1096/fj.11-189498
This study reveals that dynamic ubiquitination of histone H2B (ubH2B) governs transcriptional elongation and mRNA 3'-end processing in JAK-STAT signaling, identifying the deubiquitinase USP22 as a pivotal regulator coordinating RNA polymerase II phosphorylation and polyadenylation factor recruitment. The established H2B ubiquitination/deubiquitination cycle provides mechanistic insights into USP22's dual role in maintaining transcriptional fidelity and its potential as a therapeutic target for cancers with dysregulated STAT-dependent gene expression.
2. Whitcomb, Elizabeth A, and Allen Taylor. "Ubiquitin control of S phase: a new role for the ubiquitin conjugating enzyme, UbcH7." Cell division 4.17 (2009). https://doi.org/10.1186/1747-1028-4-17
This study identifies UbcH7 as a critical ubiquitin-conjugating enzyme regulating S-phase progression through the PTEN/Akt/Chk1 axis, revealing its dual mechanism in controlling cell cycle checkpoints and DNA replication fidelity via ubiquitin-dependent proteolysis. The findings establish UbcH7-mediated ubiquitination as a novel regulatory node coordinating proliferation dynamics and genomic stability in eukaryotic cell division.
3. Bergamaschi, Anna et al. "The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection." Journal of virology 83,10 (2009): 4854-60. https://doi.org/10.1128/JVI.00187-09
This study elucidates that the HIV-2/SIVsm accessory protein Vpx hijacks the CUL4A-DDB1 ubiquitin ligase complex via DCAF1 recruitment to degrade host restriction factors, and identifies this ubiquitin-dependent proteolytic mechanism as the molecular basis for viral replication specificity in macrophages, resolving the long-standing paradox of HIV-1 macrophage infectivity without Vpx.
4. Fryrear, Kimberly A et al. "The Sumo-targeted ubiquitin ligase RNF4 regulates the localization and function of the HTLV-1 oncoprotein Tax." Blood 119,5 (2012): 1173-81. https://doi.org/10.1182/blood-2011-06-358564
This study demonstrates that the SUMO-targeted ubiquitin ligase RNF4 mediates ubiquitination-dependent nuclear export of the HTLV-1 Tax oncoprotein, and identifies this STUbL-mediated relocalization as a novel regulatory switch that differentially modulates NF-κB and CREB signaling pathways to control viral oncogenesis.
5. Sun, Fan et al. "PDLIM2 is a novel E5 ubiquitin ligase enhancer that stabilizes ROC1 and recruits the ROC1-SCF ubiquitin ligase to ubiquitinate and degrade NF-κB RelA." Cell & bioscience 14.1 (2024):99. https://doi.org/10.1186/s13578-024-01281-x
This study reveals that the tumor suppressor PDLIM2 functions as a ubiquitin ligase enhancer (E5) by stabilizing the ROC1-SCFβ-TrCP complex to mediate nuclear RelA ubiquitination, and identifies this chromatin-bound ubiquitination cascade as a novel therapeutic target for cancers driven by persistent NF-κB activation and inflammatory pathogenesis..
Creative Biolabs: Ubiquitin Antibodies for Research
Creative Biolabs offers a broad range of antibodies designed to achieve excellent lot-to-lot consistency. For more details on our Ubiquitin antibodies, custom preparations, or technical support, contact us at info@creative-biolabs.com.
Reference
- Berk, Jason M et al. "A deubiquitylase with an unusually high-affinity ubiquitin-binding domain from the scrub typhus pathogen Orientia tsutsugamushi." Nature communications 11.1 (2020):2343. Distributed under Open Access license CC BY 4.0, without modification. https://doi.org/10.1038/s41467-020-15985-4
Anti-Ubiquitin antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-BANF1 Recombinant Antibody (3F10-4G12) (CBMAB-A0707-LY)

-
Human Anti-SARS-CoV-2 S1 Monoclonal Antibody (CBFYR-0120) (CBMAB-R0120-FY)

-
Mouse Anti-AMH Recombinant Antibody (5/6) (CBMAB-A2527-YC)

-
Mouse Anti-ELAVL4 Recombinant Antibody (6B9) (CBMAB-1132-YC)

-
Mouse Anti-APOA1 Monoclonal Antibody (CBFYR0637) (CBMAB-R0637-FY)

-
Human Anti-SARS-CoV-2 Spike Recombinant Antibody (CR3022) (CBMAB-CR014LY)

-
Mouse Anti-ADAM12 Recombinant Antibody (V2-179752) (CBMAB-A1114-YC)

-
Mouse Anti-A2M Recombinant Antibody (V2-178822) (CBMAB-A0036-YC)

-
Mouse Anti-C1QC Recombinant Antibody (CBFYC-0600) (CBMAB-C0654-FY)

-
Mouse Anti-ADGRL2 Recombinant Antibody (V2-58519) (CBMAB-L0166-YJ)

-
Mouse Anti-DES Monoclonal Antibody (440) (CBMAB-AP1857LY)

-
Mouse Anti-CA9 Recombinant Antibody (CBXC-2079) (CBMAB-C0131-CQ)

-
Mouse Anti-4-Hydroxynonenal Recombinant Antibody (V2-502280) (CBMAB-C1055-CN)

-
Mouse Anti-CECR2 Recombinant Antibody (CBWJC-2465) (CBMAB-C3533WJ)

-
Mouse Anti-AAV9 Recombinant Antibody (V2-634029) (CBMAB-AP023LY)

-
Mouse Anti-CCN2 Recombinant Antibody (CBFYC-2383) (CBMAB-C2456-FY)

-
Rat Anti-CD63 Recombinant Antibody (7G4.2E8) (CBMAB-C8725-LY)

-
Rabbit Anti-ENO2 Recombinant Antibody (BA0013) (CBMAB-0272CQ)

-
Mouse Anti-AAV-5 Recombinant Antibody (V2-503417) (CBMAB-V208-1369-FY)

-
Mouse Anti-ALB Recombinant Antibody (V2-180650) (CBMAB-A2186-YC)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot








