NPAT Antibodies
Background
NPAT gene is a nuclear protein-coding gene located in the 11q22.3 region of human chromosome. The protein encoded by NPAT is a key component of E2F transcription factor regulation pathway, which is mainly involved in the regulation process of cell cycle transition from G1 to S phase. This gene regulates the expression of histone genes by forming specific phosphorylation complexes, which is crucial for the correct assembly of chromatin during DNA replication and cell division. Studies have shown that the expression activity of the NPAT gene is directly related to the state of cell proliferation, and its dysfunction may lead to abnormal cell cycle and tumorigenesis and development. Due to its significant role in the core regulatory pathways of the cell cycle, this gene has become an important object for studying the mechanism of cell proliferation and developing cancer therapeutic targets. The research on its molecular regulatory mechanism provides key insights into the eukaryotic cell cycle regulatory network.
Structure of NPAT
NPAT is a medium-sized nucleoprotein with a molecular weight of approximately 220 kDa. This protein is composed of 1,825 amino acids, and its primary structure contains characteristic phosphorylation sites and protein-protein interaction domains. The NPAT protein functions through its N-terminal transcriptional activation domain and C-terminal nuclear localization signal, mainly located in the histone locus region within the cell nucleus. The secondary structure of this protein is mainly composed of alternating α -helices and β -folds, forming a functional interface that can interact with E2F transcription factors and cyclins. The serine residue at position 801 is a key phosphorylation site of the cyclin E-CDK2 complex, and this modification directly regulates the transcriptional activation activity of NPAT during the G1/S phase transition.
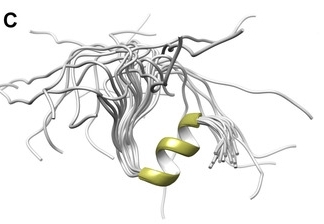 Fig. 1 The 3D high-resolution structure of the NPAT peptide in solution.1
Fig. 1 The 3D high-resolution structure of the NPAT peptide in solution.1
Key structural properties of NPAT:
- Contains multiple functional domains
- Specific phosphorylation sites to receive cell cycle signals
- Nuclear localization signals mediate its aggregation within the cell nucleus
- Protein interaction interfaces are used to assemble transcriptional complexes
Functions of NPAT
The core function of the NPAT gene is to regulate the transition of the cell cycle from the G1 phase to the S phase. In addition, it is also involved in key cellular processes such as histone biosynthesis and cell proliferation regulation.
| Function | Description |
| Cell cycle regulation | As a downstream target of the E2F transcription factor, it integrates growth signals to initiate cells into the DNA synthesis phase. |
| Histone expression regulation | In the G1 / S phase shift when activated by specific, drive histone gene transcription, provide the foundation for chromosome duplication. |
| Proliferation maintenance | Its activity is essential for continuous cell proliferation, and functional inactivation can lead to the arrest of the cell cycle at the G1 phase. |
| DNA replication coupling | Ensure that the supply of histones is synchronized with DNA replication to achieve the correct chromatin packaging of newly synthesized DNA. |
| Developmental support | In embryonic development and need a quick organization high expression of cell renewal, growth and repair of the support tissues. |
The activity of NPAT is directly regulated by the phosphorylation of the cyclin-CDK complex. This precise spatiotemporal regulation makes it a key molecular switch connecting extracellular signals with the cell cycle process.
Applications of NPAT and NPAT Antibody in Literature
1. Sang, Rong, et al. "Mxc, a Drosophila homolog of mental retardation-associated gene NPAT, maintains neural stem cell fate." Cell & Bioscience 12.1 (2022): 78. https://doi.org/10.1186/s13578-022-00820-8
Studies have shown that the NPAT gene maintains the homeostasis and differentiation of neural stem cells by ensuring the normal transcription of histones. The loss of its function will lead to DNA damage and neurogenesis abnormalities, which provides new insights into the etiology of NPAT-related intellectual disability.
2. Bucholc, Katarzyna, et al. "Structural analysis of the SANT/Myb domain of FLASH and YARP proteins and their complex with the C-terminal fragment of NPAT by NMR spectroscopy and computer simulations." International Journal of Molecular Sciences 21.15 (2020): 5268. https://doi.org/10.3390/ijms21155268
This study reveals that the FLASH and YARP proteins bind to the C-terminal of the NPAT protein in different ways through their unique domains, forming two distinctive complexes. The structure of this complex supports its binding to DNA, providing new insights into the cell cycle-dependent expression regulation mechanism of HLB assembly and histone genes.
3. Bruhn, Christopher, et al. "The Rad53CHK1/CHK2-Spt21NPAT and Tel1ATM axes couple glucose tolerance to histone dosage and subtelomeric silencing." Nature Communications 11.1 (2020): 4154. https://doi.org/10.1038/s41467-020-17961-4
Studies have shown that the DDR kinase Rad53 regulates histone dose by phosphorylating the transcription factor Spt21 (an NPAT homologous protein) to maintain metabolic homeostasis. Abnormal functions of Rad53 or Spt21 can lead to histone excess, which in turn triggers A glucose-dependent phenotype by disrupting acetyl-CoA metabolism and subtelomere silencing.
4. Xu, Shui Bo, et al. "KPNA3 regulates histone locus body formation by modulating condensation and nuclear import of NPAT." Journal of Cell Biology 224.1 (2024): e202401036. https://doi.org/10.1083/jcb.202401036
Research has found that the nuclear transport protein KPNA3 not only mediates the nuclear input of NPAT but also inhibits its C-terminal moxi-mediated self-binding through steric hindrance mechanisms, thereby effectively preventing abnormal phase separation and coagulation of NPAT in the cytoplasm and ensuring the correct assembly of histone loci bodies (HLB) within the nucleus.
5. Chen, Changwei, et al. "The genetic association of the transcription factor NPAT with glycemic response to metformin involves regulation of fuel selection." PloS one 16.7 (2021): e0253533. https://doi.org/10.1371/journal.pone.0253533
Research has found that genome-wide association studies have revealed that variations in the NPAT gene are associated with the efficacy of metformin. Animal experiments have confirmed that the absence of one npat allele weakens the regulation of metformin on respiratory exchange rate, suggesting that NPAT may mediate part of the mechanism of action of metformin by influencing the cellular fuel utilization pathway.
Creative Biolabs: NPAT Antibodies for Research
Creative Biolabs specializes in the production of high-quality NPAT antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom NPAT Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our NPAT antibodies, custom preparations, or technical support, contact us at email.
Reference
- Bucholc, Katarzyna, et al. "Structural analysis of the SANT/Myb domain of FLASH and YARP proteins and their complex with the C-terminal fragment of NPAT by NMR spectroscopy and computer simulations." International Journal of Molecular Sciences 21.15 (2020): 5268. https://doi.org/10.3390/ijms21155268
Anti-NPAT antibodies
 Loading...
Loading...
Hot products 
-
Rabbit Anti-ATF4 Recombinant Antibody (D4B8) (CBMAB-A3872-YC)

-
Mouse Anti-ENO2 Recombinant Antibody (H14) (CBMAB-E1341-FY)

-
Mouse Anti-CD24 Recombinant Antibody (SN3) (CBMAB-C1037-CQ)

-
Mouse Anti-AKT1 (Phosphorylated S473) Recombinant Antibody (V2-505430) (PTM-CBMAB-0067LY)

-
Mouse Anti-CRYAB Recombinant Antibody (A4345) (CBMAB-A4345-YC)

-
Mouse Anti-BCL2L1 Recombinant Antibody (H5) (CBMAB-1025CQ)

-
Mouse Anti-AAV8 Recombinant Antibody (V2-634028) (CBMAB-AP022LY)

-
Mouse Anti-8-oxoguanine Recombinant Antibody (V2-7719) (CBMAB-1898CQ)

-
Rat Anti-FABP3 Recombinant Antibody (CBXF-2299) (CBMAB-F1612-CQ)

-
Mouse Anti-ARID3A Antibody (A4) (CBMAB-0128-YC)

-
Mouse Anti-APOH Recombinant Antibody (4D9A4) (CBMAB-A3249-YC)

-
Mouse Anti-CORO1A Recombinant Antibody (4G10) (V2LY-1206-LY806)

-
Human Anti-SARS-CoV-2 S1 Monoclonal Antibody (CBFYR-0120) (CBMAB-R0120-FY)

-
Mouse Anti-ARG1 Recombinant Antibody (CBYCL-103) (CBMAB-L0004-YC)

-
Mouse Anti-FLI1 Recombinant Antibody (CBXF-0733) (CBMAB-F0435-CQ)

-
Mouse Anti-BHMT Recombinant Antibody (CBYY-0547) (CBMAB-0550-YY)

-
Mouse Anti-AHCYL1 Recombinant Antibody (V2-180270) (CBMAB-A1703-YC)

-
Mouse Anti-ARID1B Recombinant Antibody (KMN1) (CBMAB-A3546-YC)

-
Mouse Anti-4-Hydroxynonenal Recombinant Antibody (V2-502280) (CBMAB-C1055-CN)

-
Mouse Anti-ARSA Recombinant Antibody (CBYC-A799) (CBMAB-A3679-YC)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






