PCNA Antibodies
Background
When examining proliferating cell nuclear antigen (PCNC) at the molecular level, this trimeric ring-shaped protein reveals its conserved structure capable of encircling duplex DNA. Initially characterized in the 1980s, this essential cofactor interacts directly with DNA polymerase δ through its interdomain connecting loop. During DNA replication, PCNA functions as a sliding clamp that maintains processivity of replication machinery, while in post-replicative phases it serves as a platform for recruiting repair enzymes. Its expression pattern correlates with cell cycle progression, peaking during S phase as evidenced by nuclear immunofluorescence patterns. In oncological diagnostics, PCNA immunoreactivity demonstrates prognostic value across epithelial malignancies, particularly in identifying metastatic lesions of unknown primary origin. The protein's association with replication fork integrity has established it as a biomarker for evaluating tumor proliferative activity, while its post-translational modifications provide insights into DNA damage response pathways.
Structure of PCNA
PCNA exists as a homotrimeric sliding clamp protein, with each monomer comprising two topologically identical domains connected by an interdomain loop. The primary structure consists of 261 amino acids per subunit, featuring conserved sequences across eukaryotes. Secondary structure analysis reveals nine β-strands forming a β-sheet sandwich flanked by α-helices, creating a toroidal architecture with 35Å inner diameter.
The domains of PCNA show clear functional differentiation. The N-terminal domain (residues 1-120) and the C-terminal domain (residues 121-261) of the monomer are mainly in the β-folded conformation, and the domain connecting loop (IDCL, residues 120-136) forms the main protein interaction interface. The conserved PIP-box binding motif at residues 228-235 mediates the specific binding of DNA polymerase δ through hydrophobic interaction and electrostatic complementation mechanism.
Structural dynamics studies demonstrate that the trimer forms a closed ring through β-dimerization interfaces, with salt bridges between Glu113 and Arg110 stabilizing subunit interactions. The inner surface contains 12 positively charged residues arranged in spiral patterns, enabling nonspecific DNA backbone interactions while maintaining sliding capability.
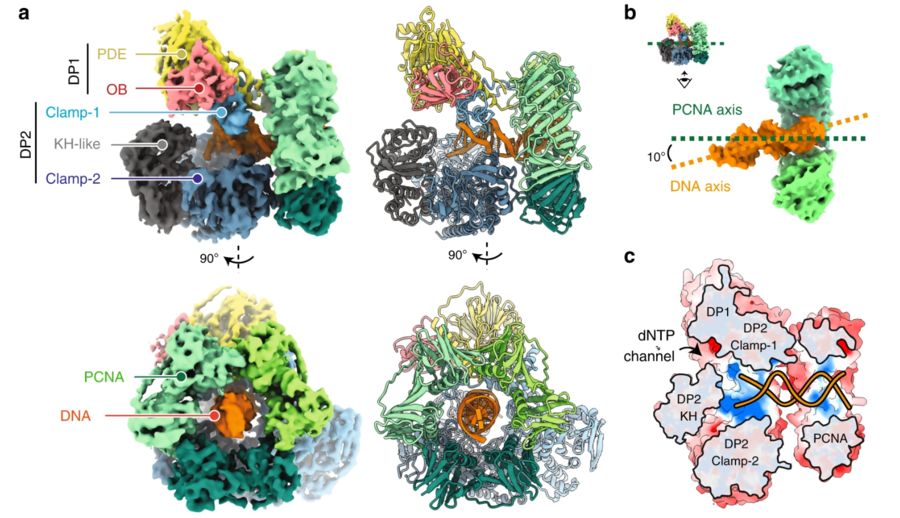 Fig. 1 Cryo-EM structure of the DNA-bound PolD–PCNA processive complex.1
Fig. 1 Cryo-EM structure of the DNA-bound PolD–PCNA processive complex.1
Functions of PCNA
As a pivotal coordinator of genomic stability, PCNA executes multifunctional regulation through its structural plasticity and dynamic interactions. This trimeric sliding clamp primarily ensures DNA replication fidelity by forming processive complexes with DNA polymerase δ/ε, where its toroidal structure encircles duplex DNA to prevent polymerase dissociation during leading-strand synthesis. Beyond replication, PCNA undergoes functional switching to orchestrate post-replicative repair: its conserved interdomain connecting loop recruits mismatch repair (MMR) proteins through PIP-box interactions, while SUMOylation at Lys110 redirects binding preference toward homologous recombination mediators.
PCNA participates in epigenetic regulation by specifically binding to chromatin modification complexes. During DNA replication, this protein works in concert with histone chaperones CAF-1 and ASF1 to mediate replication-coupled nucleosome assembly, thereby ensuring the stable transmission of chromatin structure during cell division. Phosphorylation modification at the Ser121 site can dynamically regulate its binding strength with cell cycle checkpoint kinases, forming an integrated signaling network for monitoring replication progress and regulating G2/M phase transition.
Clinical observations reveal PCNA's overexpression in 60-80% of solid tumors, correlating with enhanced replicative stress tolerance. Current therapeutic strategies exploit its functional duality: small molecules targeting the PIP-box interface disrupt replication fork progression, while ubiquitination-mimetic compounds preferentially destabilize cancer cells by hijacking error-prone repair pathways. These mechanisms position PCNA as both a diagnostic marker and a promising therapeutic node in precision oncology.
Applications of PCNA and PCNA Antibody in Literature
1. Mortusewicz, Oliver, and Heinrich Leonhardt. "XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions." BMC molecular biology 8.81 (2007). https://doi.org/10.1186/1471-2199-8-81
This study establishes PCNA as a critical orchestrator in DNA damage response coordination, demonstrating its distinct kinetic signature compared to XRCC1. Live-cell imaging revealed PCNA's stable retention at repair sites versus XRCC1's dynamic redistribution, with successive DNA damage events depleting PCNA's nuclear reservoir while maintaining XRCC1's adaptive recruitment capacity.
2. Brun, J., Chiu, R., Lockhart, K., Xiao, W., Wouters, B. G., & Gray, D. A. hMMS2 serves a redundant role in human PCNA polyubiquitination. BMC molecular biology 9, 24 (2008). https://doi.org/10.1186/1471-2199-9-24
This study elucidates PCNA's pivotal role in directing DNA damage tolerance pathways through its differential ubiquitination, and reveals compensatory mechanisms maintaining polyubiquitination in hMMS2-deficient systems, suggesting unexplored redundancy in human error-free repair processes.
3. Wang, Wan et al. "KAP1 phosphorylation promotes the survival of neural stem cells after ischemia/reperfusion by maintaining the stability of PCNA." Stem cell research & therapy 13,1 (2022):290. https://doi.org/10.1186/s13287-022-02962-5
This study demonstrates that KAP1 phosphorylation at Ser824 modulates PCNA stabilization through competitive binding interactions in cerebral ischemia/reperfusion injury, and highlights its therapeutic potential for enhancing endogenous neural stem cell proliferation by disrupting CUL4A-mediated ubiquitination pathways.
4. Ma, Rulan et al. "Downregulation of the FBXO43 gene inhibits tumor growth in human breast cancer by limiting its interaction with PCNA." Journal of translational medicine 19.1 (2021):425. https://doi.org/10.1186/s12967-021-03100-0
This study demonstrates that FBXO43 drives breast carcinogenesis through PCNA-dependent proliferative mechanisms, and highlights its therapeutic vulnerability as disrupting FBXO43-PCNA interaction effectively suppresses tumor growth while PCNA restoration reverses oncogenic phenotypes.
5. A, Peng et al. "EZH2 promotes DNA replication by stabilizing interaction of POLδ and PCNA via methylation-mediated PCNA trimerization." Epigenetics & chromatin 11.1 (2018):44. https://doi.org/10.1186/s13072-018-0213-1
This study elucidates PCNA's essential role in DNA replication through trimer stabilization, and identifies EZH2-mediated dimethylation at lysine 110 as a novel regulatory mechanism that orchestrates genomic duplication by enhancing PCNA-polymerase δ interaction.
Creative Biolabs: PCNA Antibodies for Research
Specializing in precision-engineered PCNA protein systems, Creative Biolabs deliver structural integrity-optimized complexes for diverse applications. For more details on our PCNA antibodies, custom preparations, or technical support, contact us at info@creative-biolabs.com.
Reference
- Madru, Clément et al. "Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA." Nature communications 11.1 (2023):1591. https://doi.org/10.1038/s41467-020-15392-9
Anti-PCNA antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-ADGRE2 Recombinant Antibody (V2-261270) (CBMAB-C0813-LY)

-
Mouse Anti-BIRC3 Recombinant Antibody (315304) (CBMAB-1214-CN)

-
Rabbit Anti-DLK1 Recombinant Antibody (9D8) (CBMAB-D1061-YC)

-
Mouse Anti-FeLV g27 Recombinant Antibody (1) (CBMAB-V208-1714-FY)

-
Mouse Anti-HTLV-1 gp46 Recombinant Antibody (CBMW-H1006) (CBMAB-V208-1154-FY)

-
Mouse Anti-BCL2L1 Recombinant Antibody (H5) (CBMAB-1025CQ)

-
Mouse Anti-CD24 Recombinant Antibody (2Q1282) (CBMAB-C1624-CN)

-
Rat Anti-EMCN Recombinant Antibody (28) (CBMAB-E0280-FY)

-
Mouse Anti-ACO2 Recombinant Antibody (V2-179329) (CBMAB-A0627-YC)

-
Mouse Anti-Acetyl-α-Tubulin (Lys40) Recombinant Antibody (V2-623485) (CBMAB-CP2897-LY)

-
Mouse Anti-BIRC5 Recombinant Antibody (6E4) (CBMAB-CP2646-LY)

-
Mouse Anti-14-3-3 Pan Recombinant Antibody (V2-9272) (CBMAB-1181-LY)

-
Rabbit Anti-BAD (Phospho-Ser136) Recombinant Antibody (CAP219) (CBMAB-AP536LY)

-
Mouse Anti-APP Recombinant Antibody (5C2A1) (CBMAB-A3314-YC)

-
Mouse Anti-CD2AP Recombinant Antibody (BR083) (CBMAB-BR083LY)

-
Mouse Anti-AAV8 Recombinant Antibody (V2-634028) (CBMAB-AP022LY)

-
Mouse Anti-DMD Recombinant Antibody (D1190) (CBMAB-D1190-YC)

-
Mouse Anti-ALDOA Recombinant Antibody (A2) (CBMAB-A2316-YC)

-
Rabbit Anti-AKT3 Recombinant Antibody (V2-12567) (CBMAB-1057-CN)

-
Mouse Anti-ENO2 Recombinant Antibody (H14) (CBMAB-E1341-FY)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot








