Immunoprecipitation (IP)
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sample containing many thousands of different proteins.
Individual Protein IP
Individual protein IP uses an antibody to isolate a selected protein of interest from cell lysate. The antibody binds to the protein and the antibody-antigen complex is pulled out of the sample using protein A/G-coupled agarose or magnetic beads. The beads are washed and the protein is eluted. Purified antigen obtained by IP is verified by a variety of molecular techniques such as Enzyme-linked Immunosorbent Assay (ELISA) and Western blot (WB). Our Considerations Before Starting Your IP Experiment include the following contents:
- Method Format Choice
- Binding Proteins & Beads
- Choosing the Right Primary Antibody (Antibody subtype and Binding Ability)
- Isotype & Negative Control
Refer to Protocol of Immunoprecipitation (IP) and Troubleshooting of Immunoprecipitation (IP) for more detail.
Co-Immunoprecipitation (Co-IP)
Co-IP is a powerful tool used to analyze protein-protein interactions. The main purpose of Co-IP is the identification of interaction partners to the protein of interest, such as ligands, receptors, co-factors. The basic Co-IP protocol is the same as that described for IP, and indeed any system designed for IP should also work for Co-IP. There are a number of additional factors to consider. For example, optimization of binding and wash conditions must include consideration of effects on bait-prey interactions as well as on that between antibody and bait.
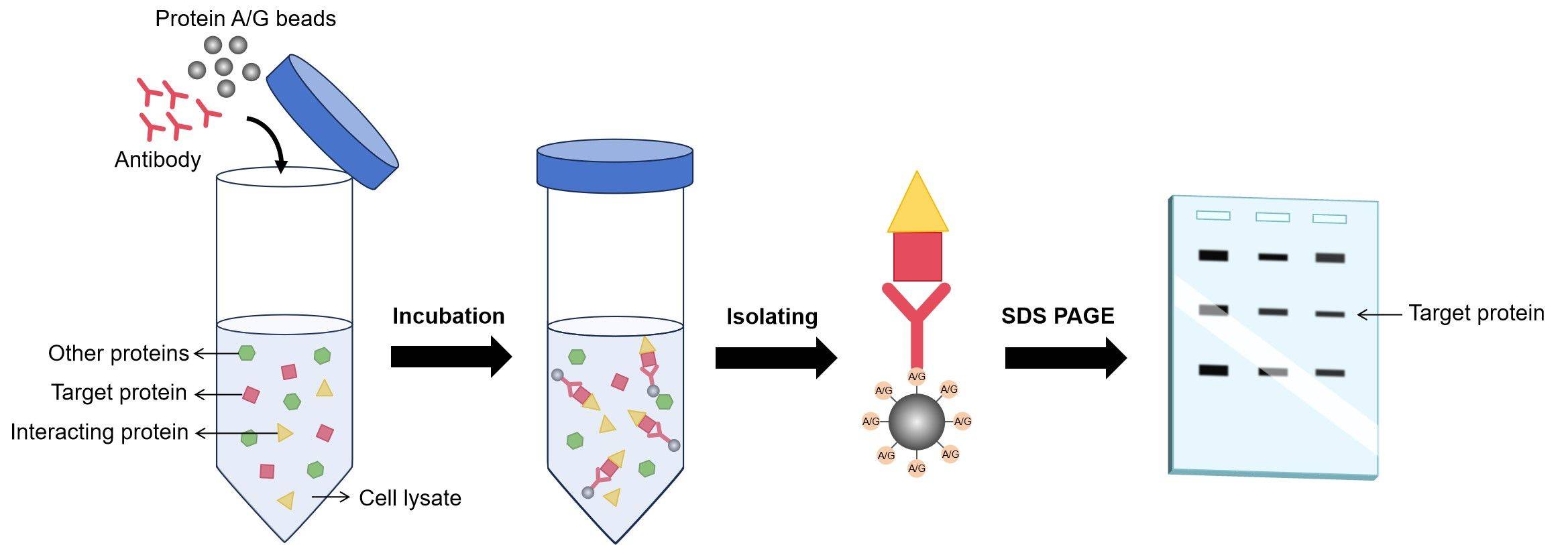 Fig.1 Scheme of the Co-IP.
Fig.1 Scheme of the Co-IP.
Chromatin Immunoprecipitation (ChIP)
ChIP is a type of IP used to investigate regions of the genome associated with a target DNA-binding protein such as transcription factors and histones or conversely to identify specific proteins associated with a particular region of the genome. It is commonly used in epigenetics research. Our Considerations Before Starting Your ChIP Experiment lists the main points to consider before starting the ChIP experiment. Refer to Protocol of Chromatin Immunoprecipitation (ChIP) and Troubleshooting of Chromatin Immunoprecipitation (ChIP) for more detail.
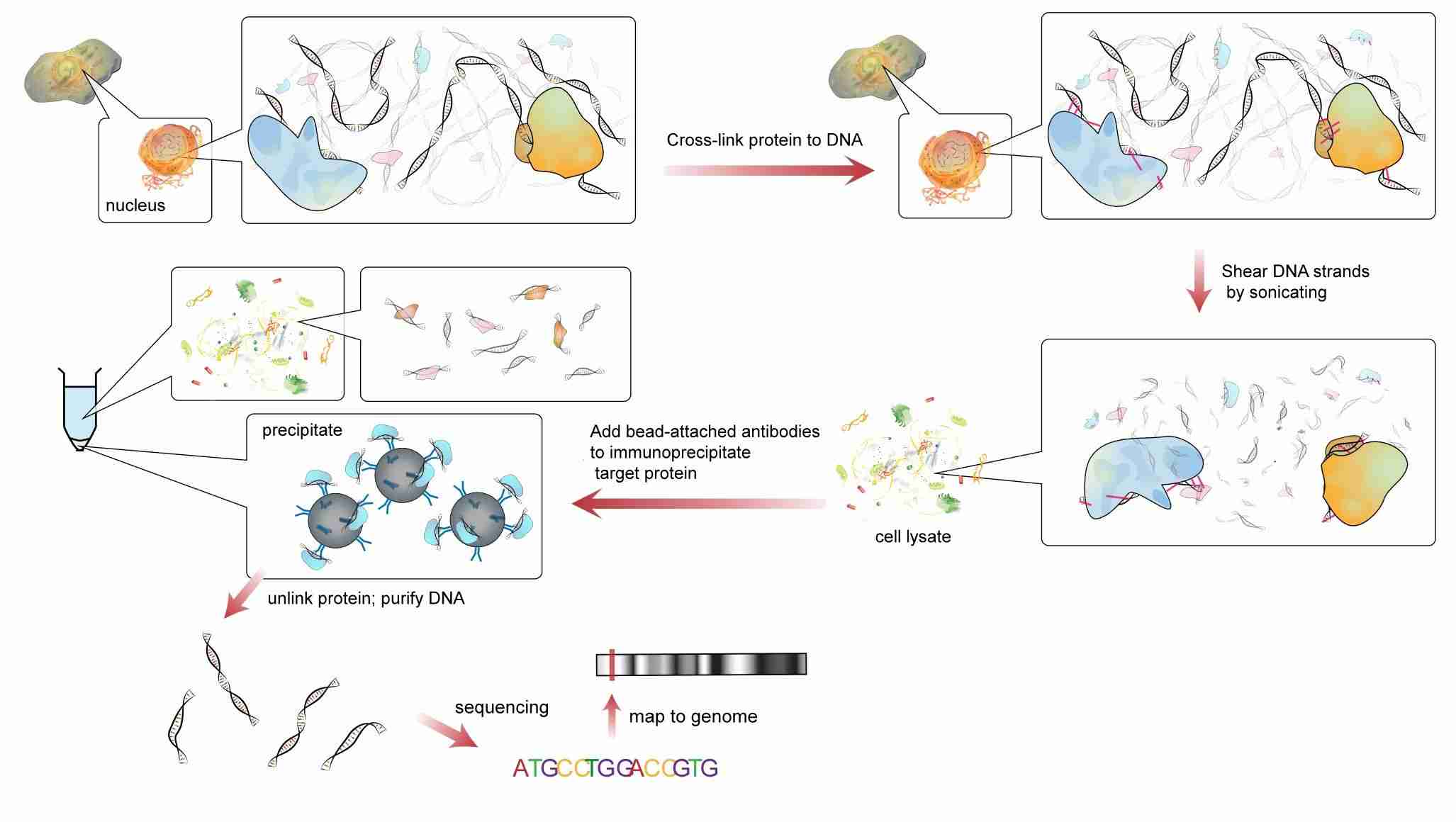 Fig.2 Scheme of the ChIP.1
Fig.2 Scheme of the ChIP.1
RNA Immunoprecipitation (RIP)
RIP is similar to ChIP but RIP is performed using an antibody that targets a specific RNA-binding protein. The RNA-protein complexes are separated by RNA extraction. Some variants of RIP, such as photoactivatable ribonucleotide-enhanced crosslinking and immunoprecipitation (PAR-CLIP) include cross-linking steps, which then require less careful lysis conditions. Immunoprecipitated RNAs can then be identified by RT-PCR and cDNA sequencing.
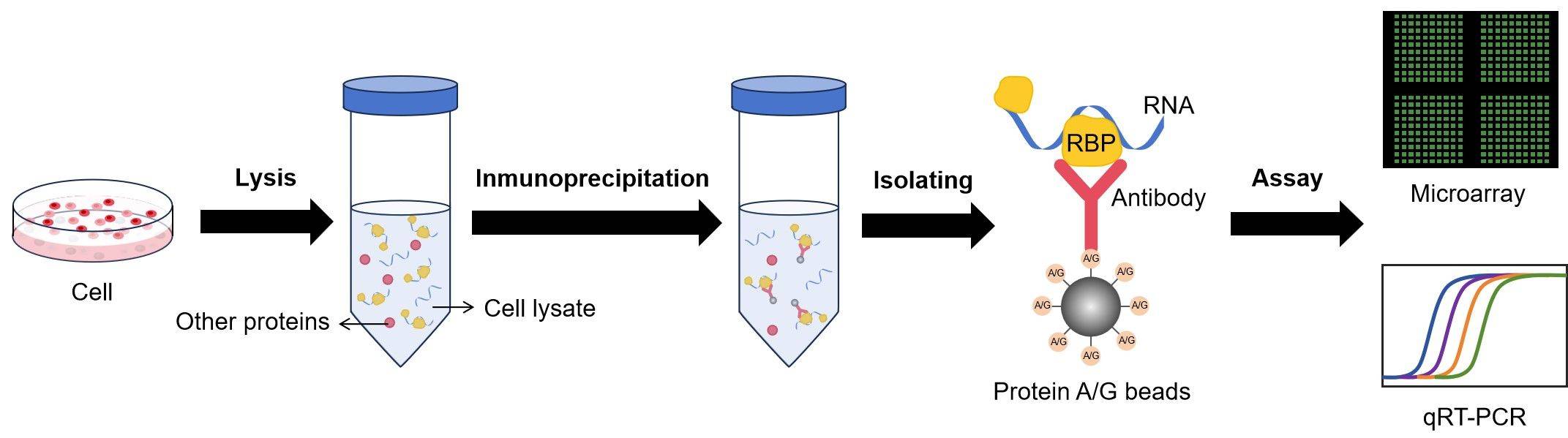 Fig.3 Scheme of the RIP.
Fig.3 Scheme of the RIP.
Tagged Protein IP
Due to the lack of an available antibody for a specific target, many proteins cannot be immunoprecipitated. To circumvent this problem, proteins can be tagged with an epitope to which a high-affinity antibody is available and ectopically expressed in the cell of interest. These tags can be either short peptide sequences or fluorescent proteins, including:
- FLAG-peptide sequence DYKDDDDK
- c-Myc-peptide sequence EQKLISEEDL
- Hemagglutinin (HA)-peptide sequence YPYDVPDYA
- V5-peptide sequence GKPIPNPLLGLDST
- Green fluorescent protein (GFP)
Immunoprecipitation Methodology
Target antigens are usually immunoprecipitated from complex solutions, such as cell lysates, the goal being to isolate and eventually detect and measure a specific protein. The basic protocol for performing an IP is summarized in Protocol of Immunoprecipitation (IP), where the order of steps can be done in two different ways. In one sequence, the antibody against the target is pre-immobilized onto an insoluble support, such as agarose or magnetic beads, and then incubated with a cell lysate containing the target protein. Alternatively, free antibody is allowed to form immune complexes in the lysate and then the complexes are retrieved by the beads.
Applications
- Identify the activation status of proteins
- Determine post-translational protein modifications
- Measure the molecular weight of a given protein
- Capture protein-binding molecules in the study of protein-protein and protein-nucleic acid interactions
Advantages of IP
- IP is ideal for small-scale enrichment of proteins.
- IP is fast and relatively easy in comparison to affinity chromatography which is time-consuming and involves cycles of bind and washing.
- Co-IP is a powerful technique to identify protein interactions with physiological importance, since both bait and its prey proteins are in their native conformations during the whole procedure. Thus, co-IP is considered as the golden standard assay for protein-protein interaction.
Limitations of IP
- Co-IP might not be able to capture low affinity and transient protein interactions.
- The protein-protein interaction detected by co-IP may be indirect, because a third protein might be sandwiched in between the bait and prey proteins. As a result, results from co-IP must be further validated by other assays.
- Co-IP is limited by the availability of antibodies that recognize the bait protein. In some cases, the antibody that recognizes the bait protein might bind to the interacting site between bait and prey proteins and interfere or disrupt the interaction.
- Co-IP can only verify protein interactions between suspected interaction partners, thus it is not suitable for high-throughput screening approach.
Do you have any suggestions on how to perform a successful IP experiments? Do you have questions on a particular step in the protocol? Find what you need from the quick links below.
- Immunoprecipitation (IP) Protocols & Troubleshooting & Guide
- Protocol of Immunoprecipitation (IP)
- Troubleshooting of Immunoprecipitation (IP)
- Chromatin Immunoprecipitation (ChIP)
- Protocol of Chromatin Immunoprecipitation (ChIP)
- Troubleshooting of Chromatin Immunoprecipitation (ChIP)
Reference
- From Wikipedia: Jkwchui derivative work. This file was derived from: Chromatin immunoprecipitation sequencing.svg, CC BY-SA 3.0, https://commons.wikimedia.org/wiki/File:Chromatin_immunoprecipitation_sequencing.svg
