Post-Translational Modification (PTM)
As a world leader in antibody development, Creative Biolabs strives to offer the best of antibody production to its customers. Our desire is to connect the most ambitious and innovative antibody production projects with our highly skilled and enthusiastic team of experts. Our expertise and state-of-the-art technology, coupled with the passion we put into your projects every day, explain our unparalleled track record.
Protein post-translational modifications (PTMs) increase the functional diversity of proteome by covalently adding functional groups or proteins, proteolysis of regulatory subunits or degradation of the full protein. Generally, various PTMs have been found including phosphorylation, glycosylation, ubiquitination, nitrosylation, methylation, acetylation and lipidation. These PTMs almost affect all aspects of normal cell biology and pathogenesis, such as signal transduction, development, cell structure, mitosis, DNA modification, cancer, neurodegeneration, etc. Thus, it is essential to identify and understand PTMs during the study of cell biology and treatment and prevention for disease.
Introduction of PTM
PTM is a process that occurs to one or more amino acids on a protein after the protein has been translated by a ribosome. Proteins are usually produced by ribosomes that translate mRNA into polypeptide chains and then form mature protein products via PTM. PTMs play an important role in regulating protein folding, targeting specific subcellular compartments, interacting with ligands or other proteins, and in signal transduction pathways.
PTM can occur on the amino acid side chains or at the protein's C- or N- termini. Phosphorylation is one of the most common PTMs and the mechanism is associated with the activity regulating of enzymes. Glycosylation is a PTM process presenting in many eukaryotic and prokaryotic proteins that carbohydrate molecules can attach to them, which can improve protein folding and stability as well as regulate protein function. Lipidation is another PTM form that lipid molecules are linked to a protein or part of a protein attached to the cell membrane.
Other PTMs result in the cleaving peptide bonds, the maturing of a propeptide by removing the initiator methionine residue, the formation of disulfide bonds from cysteine residues. Some types of post-translational modification can regulate the oxidative stress. For example, Carbonylation targets the protein causing the modified degradation and resulting in the formation of protein aggregates. Therefore, specific PTMs of amino acid can be considered as biomarkers to indicate oxidative damage.
Currently, PTM of proteins can be detected by various techniques, such as mass spectrometry, Eastern blotting, Western blotting and so on.
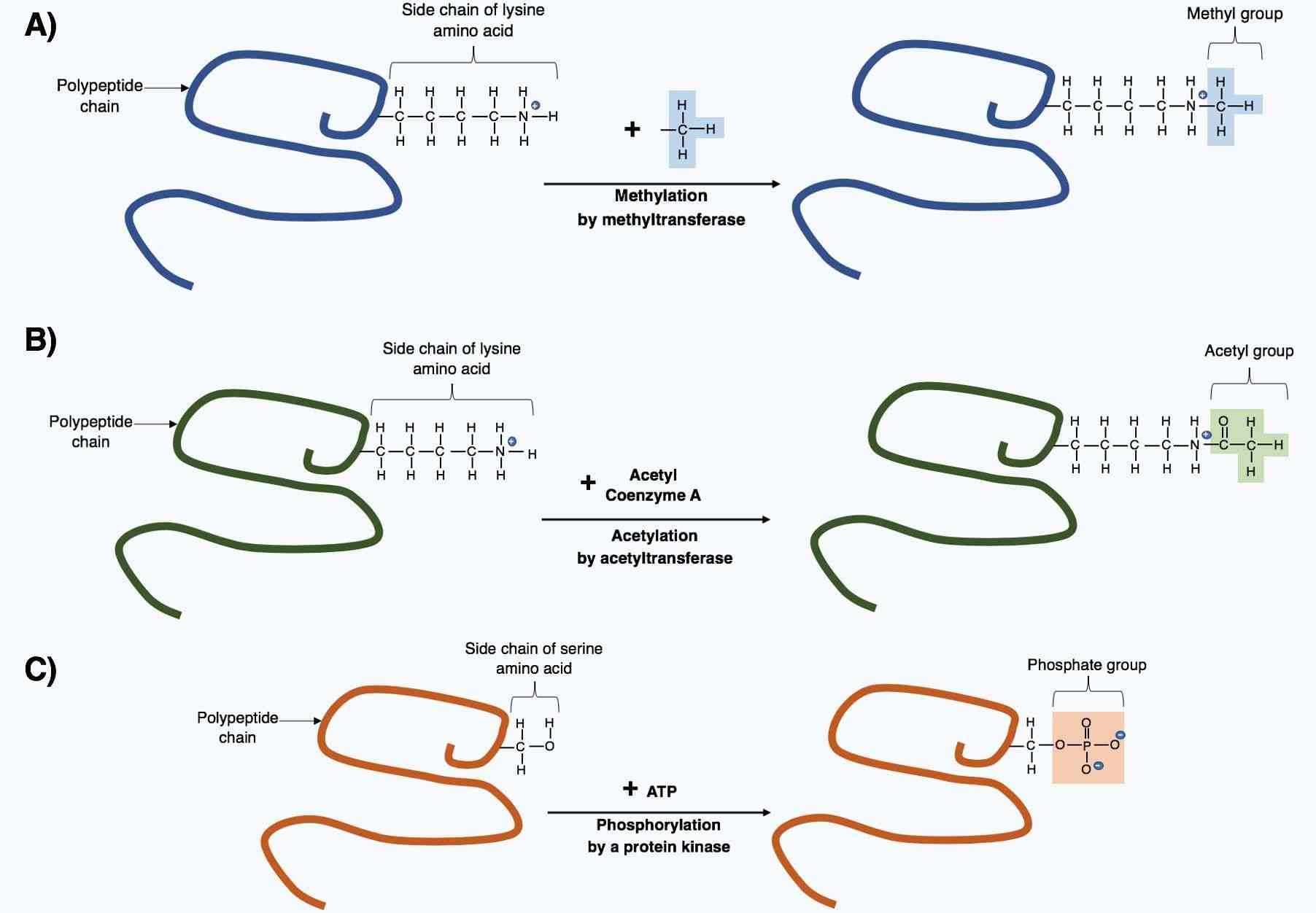 Fig.1 The post-translational modification of polypeptides.1
Fig.1 The post-translational modification of polypeptides.1
Types of PTM
To date, a variety of PTMs have been identified and understood. This overview only presents several of the most common types of PTMs in protein research today including phosphorylation, glycosylation, ubiquitination, S-nitrosylation, Methylation, N-acetylation and Lipidation.
- Phosphorylation
Phosphorylation is one of the most important and well-studied PTMs. The process is a molecule attached to a phosphoryl group of serine, threonine or tyrosine residues. Phosphorylation and its counterpart dephosphorylation involve in many cellular processes in biology, including cell cycle, growth, apoptosis and signal transduction pathways. For example, the modification can activate or deactivate almost half of the enzymes of yeast. Many proteins in eukaryotes are phosphorylated temporarily.
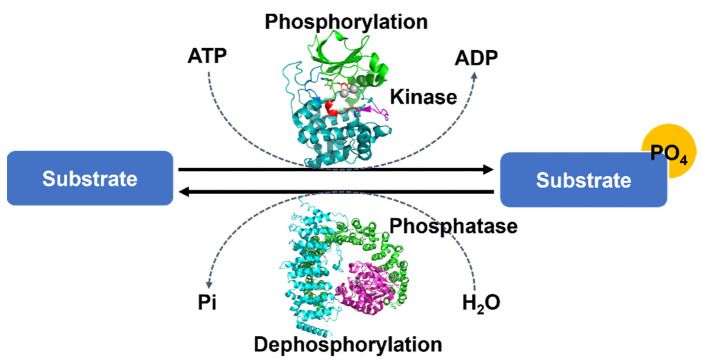 Fig.2 The overall mechanism of protein phosphorylation.2,7
Fig.2 The overall mechanism of protein phosphorylation.2,7
- Glycosylation
Glycosylation is the reaction in which carbohydrate is linked to a hydroxyl or other functional group of another molecule. In biology, glycosylation attaches glycans to proteins or other organic molecules, and mainly influences the enzymatic process. Glycosylation represents one of the major post-translational modifications that can significantly regulate protein folding, conformation, distribution, stability and activity. Glycans play a critical role in membrane and secreted proteins, and most of proteins synthesized in the rough endoplasmic reticulum undergo glycosylation. Glycosylation also exists in the cytoplasm and nucleus as the O-GlcNAc modification. A glycosylation is a process of engineered antibodies to remove glycosylation. In general, N-linked glycans attach to the nitrogen of asparagine or arginine side-chains. O-linked glycans attach to the hydroxyl oxygen of serine, threonine, tyrosine, hydroxylysine, etc. Phosphoglycans attach to a phosphoserine, C-linked glycans attach to a carbon of tryptophan side-chain.
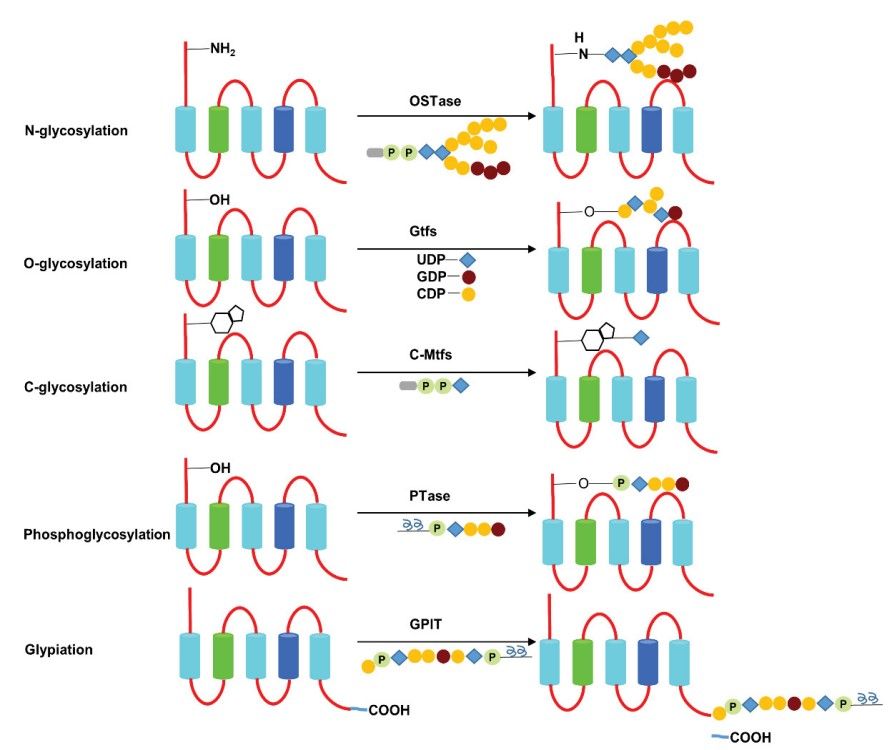 Fig.3 Classification and process of protein glycosylation.3,7
Fig.3 Classification and process of protein glycosylation.3,7
- Ubiquitination
Ubiquitin is a small 8-kDa polypeptide regulatory protein in eukaryotic organisms, and it was firstly identified in 1975. Ubiquitin is encoded by four genes in the human including UBB, UBC, UBA52 and RPS27A.
Ubiquitination (also known as ubiquitylation or ubiquitinylation) is a PTM that the addition of ubiquitin to a substrate protein. Ubiquitination regulates the protein function in many ways including marking proteins for degradation by the proteasome, altering the cellular location of protein, affecting the activity of proteins, and improving or blocking protein interactions. Ubiquitination is composed of three main steps, i.e. activation, conjugation, and ligation, which performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. These sequential cascades result in ubiquitin binding to lysine residues of proteins via an isopeptide bond, cysteine residues via a thioester bond, serine and threonine residues via an ester bond, or amino groups at the N-terminal of proteins via a peptide bond.
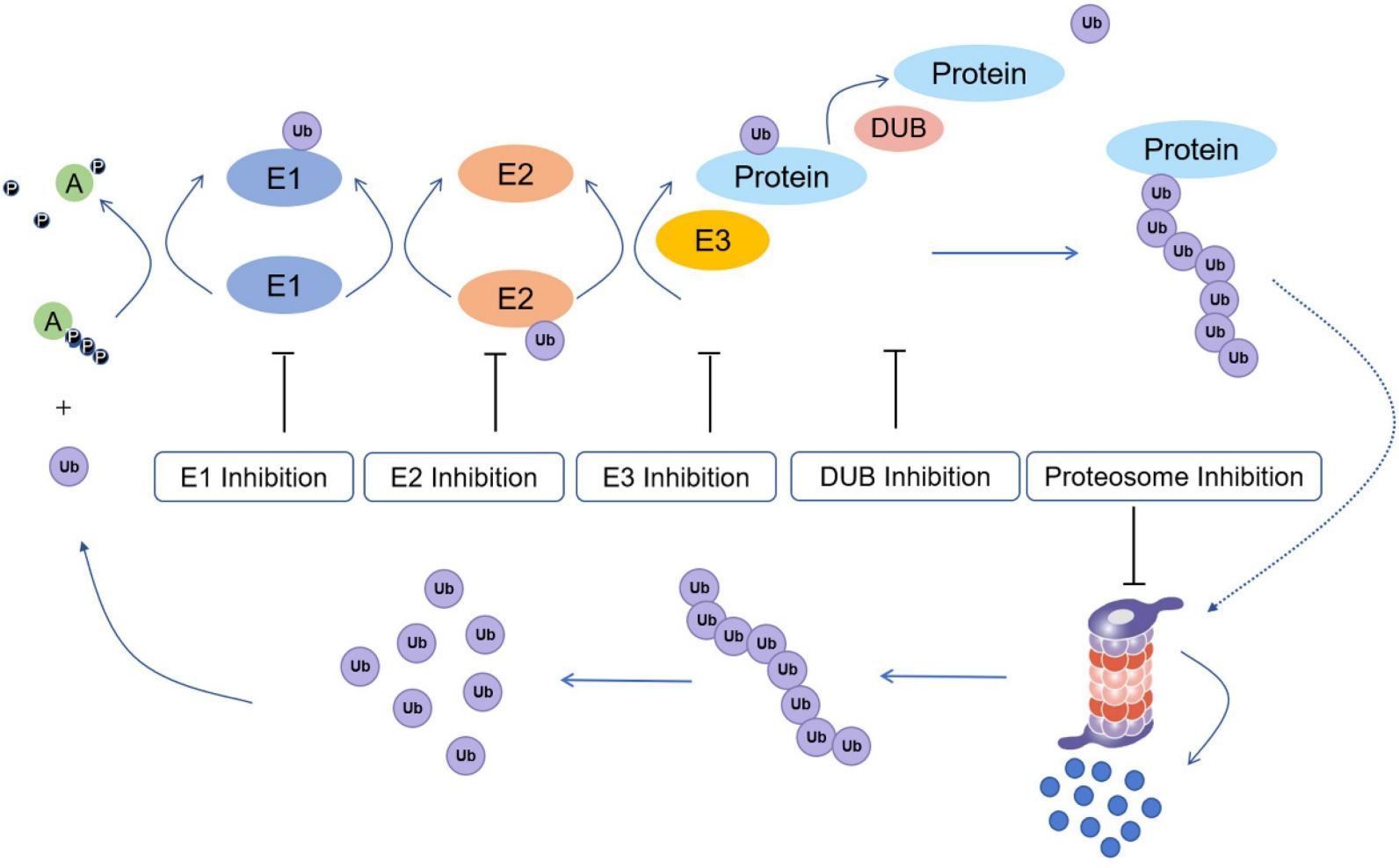 Fig.4 Ubiquitination process and potential drug inhibition targets.4,7
Fig.4 Ubiquitination process and potential drug inhibition targets.4,7
- S-nitrosylation
S-Nitrosylation is a critical PTM in which a nitric oxide group (-NO) covalently attaches to cysteine thiol within a protein to form an S-nitrosothiol (SNO). S-nitrosylation plays important roles in bacteria, yeast and plants and in all mammalian cells. It is a fundamental mechanism during cellular signaling across phylogeny and attributes to the large part of NO bioactivity.
S-nitrosylation is precisely targeted and reversible PTM to regulate a variety of cellular responses. S-nitrosylation often depends on enzymatic activity (S-nitrosylases) that facilitates the attachment of NO to proteins. The redox mechanism of S-nitrosation in biological system was also found. One example is aberrant S-nitrosylation of the NMDA receptor can result in S-nitrosylation-related diseases. S-nitrosylation also involves in the physiology and dysfunction of cardiac, airway and skeletal muscle and the immune system.
- Methylation
Methylation is a kind of PTM, which describes the transfer of methyl to a substrate, or the substitution of an atom (or group) by a methyl group. Methylation can contribute to increasing the hydrophobicity of the protein and neutralizing a negative amino acid charge when bound to carboxylic acids.
In biological systems, methylation is mediated by enzymes called methyltransferases, and S-adenosyl methionine (SAM) is the primary methyl group donor. Methylation is associated with the modification of heavy metals, regulation of gene expression, regulation of protein function, and RNA processing. Methylation is a well-known mechanism of epigenetic regulation, as histone methylation and demethylation regulates the availability of DNA for transcription. Amino acid residues can be linked to one or more methyl groups to increase the modification effect. Commonly, the N-methylation is irreversible, while the O-methylation is potentially reversible.
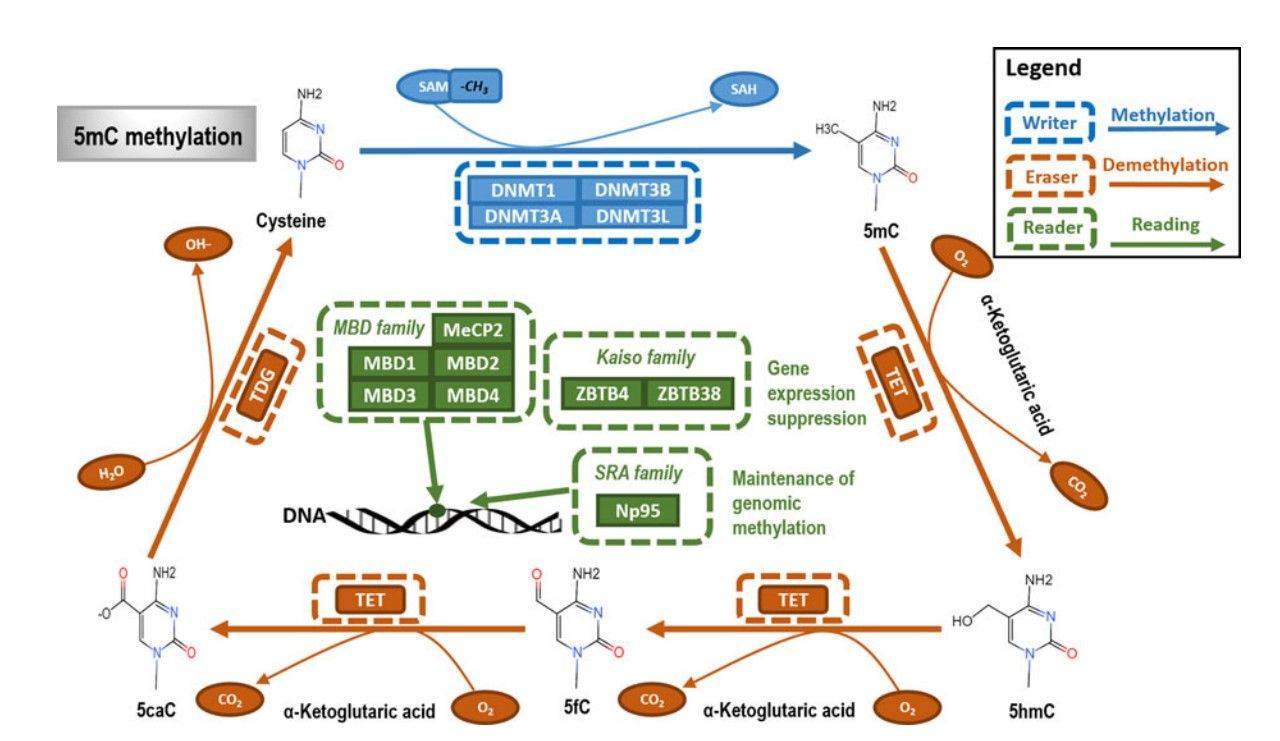 Fig.5 The primary type and mechanism of DNA methylation.5,7
Fig.5 The primary type and mechanism of DNA methylation.5,7
- N-acetylation
N-acetylation is a common PTM process in which an acetyl group can be transferred to nitrogen. It generally occurs in eukaryotic proteins through both irreversible and reversible mechanisms. N-terminal acetylation requires the cleavage of N-terminal methionine through methionine aminopeptidase (MAP), and then replaces the amino acid with an acetyl group from acetyl-CoA by N-acetyltransferase (NAT) enzymes. This type of acetylation is the main form in eukaryotic proteins.
Lysine acetylation at the ε-NH2 of lysine on histone N-termini is a common PTM to regulate the gene transcription. Histone acetylation is a reversible PTM that promotes transcription by reducing chromosomal condensation, and the acetylation of these lysine residues is regulated by transcription factors with histone acetyletransferase (HAT) activity. Sirtuins are a group of NAD-dependent deacetylases that target histones. They can maintain gene silencing by histone acetylation and genomic stability.
Besides histone acetylation, cytoplasmic protein acetylation also has been found to play a critical role in cell biology. Protein acetylation presently can be detected by chromatin immunoprecipitation (ChIP) using acetyllysine-specific antibodies or by mass spectrometry.
- Lipidation
Lipidation is a PTM to target proteins to membranes in organelles, vesicles and the plasma membrane. Lipidation has four types including C-terminal glycosyl phosphatidylinositol (GPI) anchor, N-terminal myristoylation, S-myristoylation and S-prenylation.
GPI anchors attach proteins to the plasma membrane. GPI-anchored proteins are often localized to cholesterol- and sphingolipid-rich lipid rafts, which serve as signaling platforms on the plasma membrane. This PTM is reversible and GPI anchor can be released from the protein.
N-myristoylation is a PTM to endow proteins a hydrophobic handle for membrane localization. The myristoyl group is a 14-carbon saturated fatty acid (C14), which provides the protein sufficient hydrophobicity and affinity for membranes. Thus, N-myristoylation serves as a conformational localization switch by protein conformational changes to affect the availability of the handle for membrane attachment.
S-palmitoylation can attach a C16 palmitoyl group of palmitoyl-CoA to the thiolate side chain of cysteine residues by palmitoyl acyltransferases (PATs). This anchor can permanently anchor the protein to the membrane through the longer hydrophobic group. This localization is reversible and thioesterases can break the link between the protein and the anchor. Therefore, S-palmitoylation acts as a switch to regulate membrane localization.
S-prenylation is a process of covalently linking farnesyl (C15) or geranylgeranyl (C20) groups to specific cysteine residues by farnesyl transferase (FT) or geranylgeranyl transferases (GGT I and II). S-prenylation often displays significantly hydrolytically stable. Prenylation generally creates in the ER and is a stepwise process of PTMs, which can be broken by Rce1.
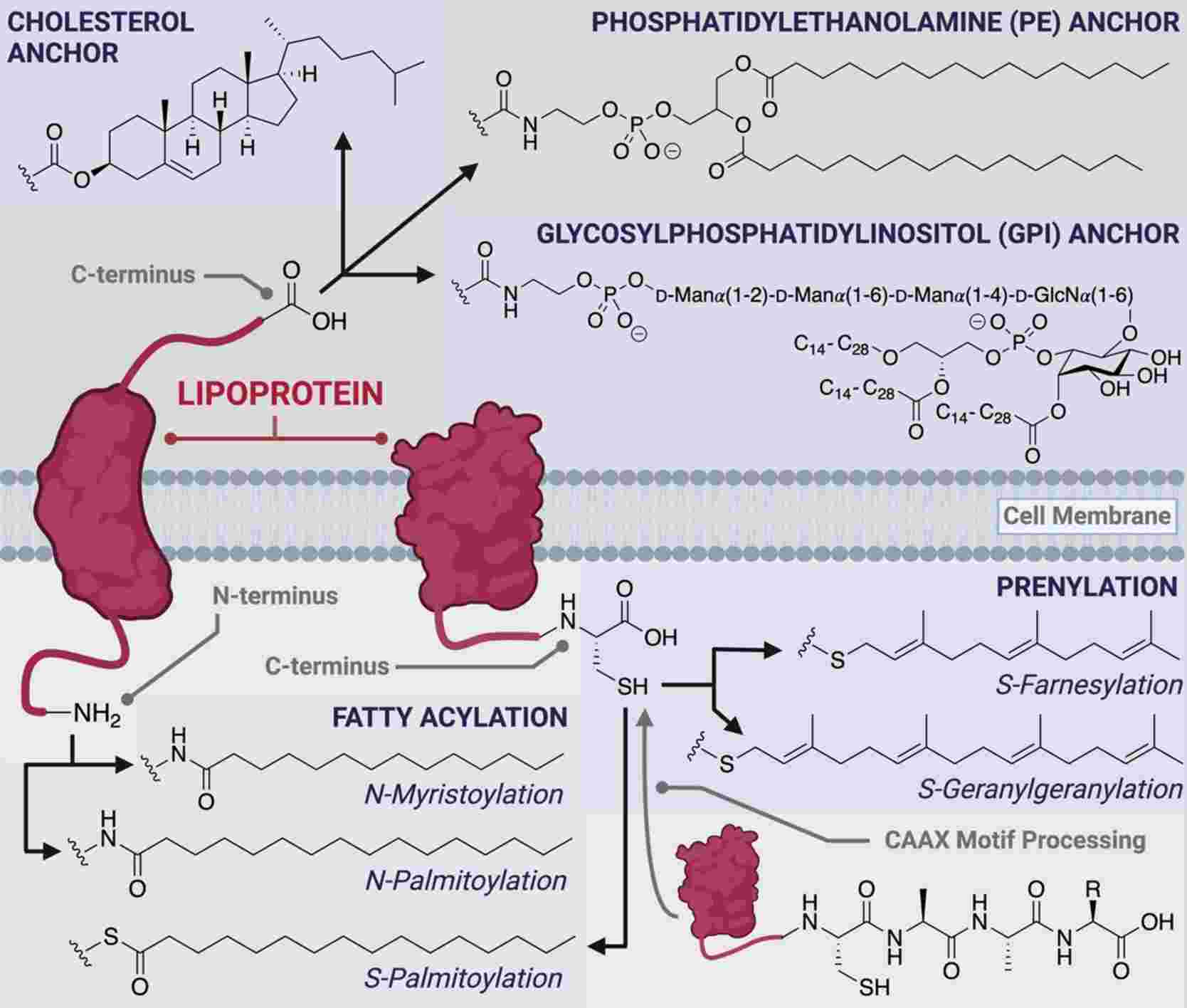 Fig.6 The different classes of co- and post-translational peptide and protein lipid modifications.6,7
Fig.6 The different classes of co- and post-translational peptide and protein lipid modifications.6,7
PTM Antibodies
PTM antibodies are used to study the spatiotemporal information of modified protein. For example, PTM antibodies can be used to study the existence or relative amounts of the modified protein by WB method, or quantitatively characterize cell populations bearing the modification by flow cytometry.
PTM antibodies can also be used to detect the protein-protein, protein-DNA and/or protein-RNA complexes containing the modified protein. One example is the Chromatin Immunoprecipitation (ChIP) used to identify genomic regions containing specific modified histone residues. It is the core application of PTM antibody in epigenetics. In addition, PTM antibodies are used in high-throughput screening of RNA interference (RNAi), to study the regulation of signaling pathways or describe the fate of a phosphorylated protein.
As a global leading antibody development company, Creative Biolabs strives not only at providing best-in-class products and services but also a broad range of solutions to carefully adapt to each project specifications. Our professional team is also willing to share its long-standing experience with its customers. If you have more questions about PTMs or PTM antibodies, please feel free to contact us.
References
- From Wikipedia: By Kep17 - Own work, CC BY-SA 4.0, without modification. https://commons.wikimedia.org/wiki/File:Post-translational_modification_through_the_addition_of_small_chemical_groups.png
- Seok, Seung-Hyeon. "Structural insights into protein regulation by phosphorylation and substrate recognition of protein kinases/phosphatases." Life 11.9 (2021): 957.
- Wang, S., et al. "N-linked Glycosylation and its Potential Application in Drug Development." Health Sci. J 14 (2020): 743.
- Song, Ying-Qi, et al. "Ubiquitination regulators discovered by virtual screening for the treatment of cancer." Frontiers in Cell and Developmental Biology 9 (2021): 665646.
- Dai, **aofeng, et al. "Methylation multiplicity and its clinical values in cancer." Expert reviews in molecular medicine 23 (2021): e2.
- Hanna, Cameron C., et al. "Chemical synthesis and semisynthesis of lipidated proteins." Angewandte Chemie International Edition 61.15 (2022): e202111266.
- Distributed under Open Access license CC BY 4.0, without modification.
