Proteogenomic
Proteogenomics is an area of research at the interface of proteomics and genomics. It was initially used to describe studies in which proteomic data are used for improved genome annotation and characterization of the protein-coding potential. Proteogenomics has since been broadened to include any type of application in which a proteogenomics-like approach is used to interpret tandem mass spectrometry (MS/MS) spectra. In this approach, customized protein sequence databases are used to help identify novel peptides from MS-based proteomic data. In turn, the proteomic data can be used to provide protein-level evidence of gene expression and to help refine gene models.
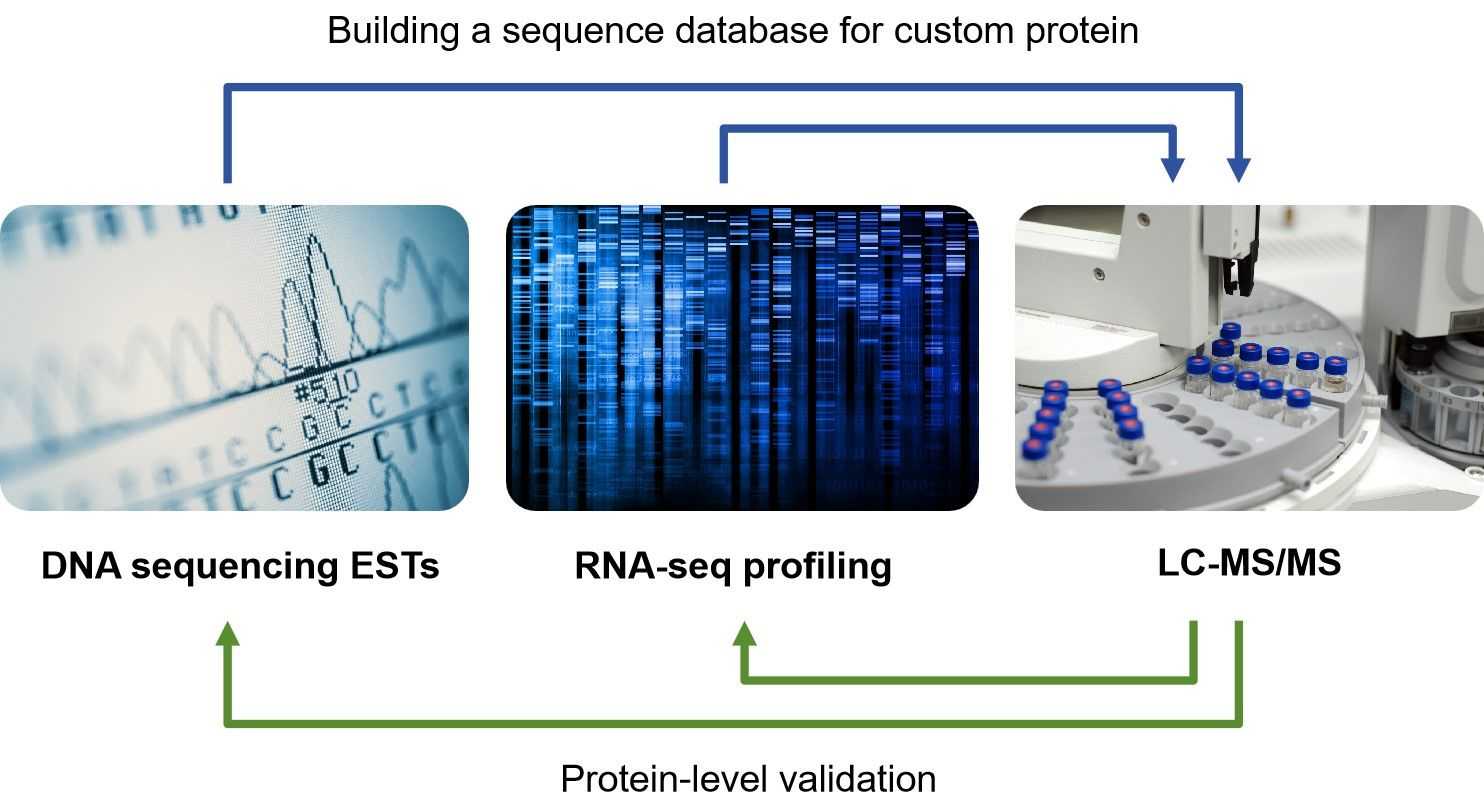 Fig.1 The concept of proteogenomics.
Fig.1 The concept of proteogenomics.
As Fig.1 shown, in a proteogenomic approach, novel peptides are identified by searching MS/MS spectra against customized protein sequence databases containing predicted novel protein sequences and sequence variants. These databases are generated using genomic and transcriptomic sequence information.
The main idea behind the proteogenomic approach is to identify peptides by comparing MS/MS data to protein databases that contain predicted protein sequences. Type of peptides identified in proteogenomics can be classified as intergenic or intragenic. Intergenic peptides map to regions located between annotated gene models, whereas intragenic peptides map to genomic regions contained within or in close proximity to an annotated gene model. The protein database is generated in a variety of ways through the utilization of genomic and transcriptomic data. Below are some of the ways in which protein databases are generated:
Generation of Customized Protein Sequence Databases
- Six-frame translation of the genome
- Ab initio gene prediction
- Expressed sequence tag (EST) data
- Annotated RNA transcripts.
- RNA-seq data
- Variant sequences
- Other specialized databases such as ECgene database, the Pseudogene.org database, the noncoding RNA sequence database.
Considerations for Peptide Identification Using Customized Protein Databases
- Balance between the completeness of the database and its size.
- Strategies for improving the sensitivity of peptide identification.
- Accurate statistical assessment of the identification confidence for different classes of peptides.
- Class-specific analysis and false discovery rates (FDR) estimation.
- False peptide identifications of nonrandom nature.
- Levels of data summarization and inference of novel events.
- Defining novel peptides.
Applications
The feasibility of various proteogenomic applications has been demonstrated in multiple studies in human and in many model organisms, including in Plasmodium falciparum, Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana and Anopheles gambiae. Recent improvements in proteomic technologies, coupled with wide availability of high-throughput DNA and transcriptome sequencing data, have led to a resurgence of proteogenomic studies.
- Discovery of novel protein-coding regions
- Computational prediction of short open reading frames and translation initiation sites
- Analysis of alternative splicing
- Identification of sequence variants
- Providing evidence of protein expression for novel gene fusions and chimeric transcripts
- Metaproteomics study
