XPA Antibodies
Background
The protein encoded by the XPA gene, as a core component of the nucleotide excision repair (NER) complex, mainly participates in identifying and repairing damage sites on the DNA double helix. This protein ensures that cells can maintain genomic stability under the influence of ultraviolet radiation or chemical mutagens by working in synergy with multiple repair enzymes. Since its first identification in 1990, mutations in the XPA gene have been confirmed to be directly related to Xeroderma Pigmentosum, a human genetic disorder, in which patients exhibit extreme UV sensitivity and cancer susceptibility. Its highly conserved domain and specific DNA binding ability have become a classic model for studying the mechanism of DNA damage repair, significantly promoting the progress in the fields of cancer treatment and targeted drug development for DNA repair.
Structure of XPA
XPA is a zinc finger protein with a molecular weight of approximately 31.3 kDa, and its size is highly conserved across different species. The following is a comparison of the molecular weights of XPA proteins in different mammals:
| Species | Human | Mouse | Rat | Bovine |
| Molecular Weight (kDa) | 31.3 | 31.2 | 31.3 | 31.1 |
| Primary Structural Differences | Zinc-containing domain | Highly conserved zinc finger structure | 93% homologous to humans | Lack of C-terminal polymorphism |
This protein is composed of 273 amino acids and contains a central zinc finger domain (amino acids 98-219) and two functional domains. The N-terminal domain (amino acids 1-97) is responsible for binding to the RPA32 protein subunit, while the C-terminal domain (amino acids 220-273) mediates the interaction with the TFIIH complex. XPA specifically recognizes DNA double helix twisted damage through its zinc finger domain, where conserved amino acid residues such as Phe129 and Arg133 directly participate in DNA binding. The secondary structure of proteins is mainly composed of α -helices and β -folds, forming a typical zinc finger motif, which stabilizes the three-dimensional structure through zinc ions. This structural feature enables it to serve as a molecular scaffold in nucleotide excision repair, precisely guiding repair factors to the injury site.
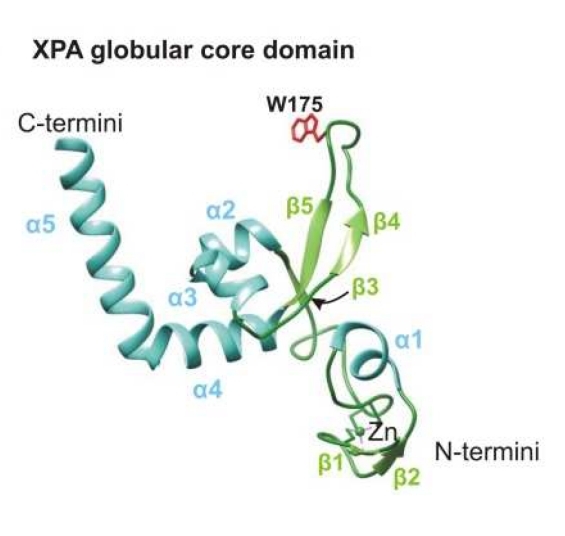 Fig. 1 A structural model of the XPA globular core domain.1
Fig. 1 A structural model of the XPA globular core domain.1
Key structural properties of XPA:
- Contains a central zinc finger domain (ZF domain)
- With two functional structure domain: N and RPA protein interactions, C side is responsible for the recruiting TFIIH repair factor, etc
- Conserved histidine and cysteine residues chelate zinc ions
- Form hydrophobic core region, safeguard its with damaged DNA and other nucleotide excision repair (NER) the effective combination of complex components
Functions of XPA
The core function of the XPA gene is to participate in the identification and repair of DNA damage. Its main functions include:
| Function | Description |
| DNA damage identification | Specifically identify DNA double helix twisting damage caused by ultraviolet rays or chemical substances as the initial sensor for nucleotide excision repair (NER). |
| Repair complex assembly | Recruit key repair proteins such as RPA, TFIIH, and XPF to the injury site to form a functional repair complex. |
| DNA damage verification | Verify the type of damage and ensure the specificity of the repair to prevent normal DNA from being wrongly excised. |
| Maintenance of genomic stability | By efficiently repairing UV-induced damage such as pyrimidine dimers, it prevents mutation accumulation and cell carcinogenesis. |
| Cell cycle regulation is involved | Plays a role in DNA damage checkpoints that delay the cell cycle so that repair can be done. |
The binding of XPA protein to DNA does not have enzymatic activity, but it exerts a molecular scaffold function through protein-protein and protein-DNA interactions. Its repair activity strictly depends on the heterodimer formed with replication protein A (RPA), and this cooperative binding enhances the affinity and specificity of XPA for damaged DNA. Unlike the broad substrate recognition characteristics of other DNA repair proteins, XPA specifically targets helical twisted damage, demonstrating its precise regulatory role in maintaining genomic integrity.
Applications of XPA and XPA Antibody in Literature
1. Krasikova, Yuliya S., Olga I. Lavrik, and Nadejda I. Rechkunova. "The XPA Protein—Life under Precise Control." Cells 11.23 (2022): 3723. https://doi.org/10.3390/cells11233723
The article indicates that the XPA protein is a key scaffold protein in nucleotide excision repair (NER), responsible for interacting with multiple repair factors and assembling repair complexes. Its activity and expression are precisely regulated by various post-translational modifications.
2. Borszéková Pulzová, Lucia, Thomas A. Ward, and Miroslav Chovanec. "XPA: DNA repair protein of significant clinical importance." International journal of molecular sciences 21.6 (2020): 2182. https://doi.org/10.3390/ijms21062182
The article indicates that the XPA protein is a core scaffold protein for nucleotide excision repair (NER) and plays a key role in platinum-based chemotherapy resistance. Its expression level is related to the therapeutic response. It mediates damage verification and repair complex assembly through protein interaction and is a potential tumor prognostic marker and therapeutic target.
3. Fadda, Elisa. "Role of the XPA protein in the NER pathway: A perspective on the function of structural disorder in macromolecular assembly." Computational and structural biotechnology journal 14 (2016): 78-85. https://doi.org/10.1016/j.csbj.2015.11.007
The article indicates that XPA proteins assemble the NER complex in a "beading" pattern through their intrinsic disordered regions as key scaffolds. This flexible structure effectively avoids the steric hindrance of DNA expansion bubbles and regulates the repair efficiency by modularly interacting and orderly coordinating the binding of repair factors.
4. Krasikova, Yuliya S., et al. "Does the XPA–FEN1 Interaction Concern to Nucleotide Excision Repair or Beyond?." Biomolecules 14.7 (2024): 814. https://doi.org/10.3390/biom14070814
The article indicates that XPA, as a core scaffold protein of NER, can form a complex with FEN1 and moderately inhibit its activity. Studies have shown that XPA can form a ternary complex together with RPA and FEN1, which may be involved in regulating NER repair synthesis and other DNA metabolic processes.
5. Kose, Cansu, et al. "Cross-species investigation into the requirement of XPA for nucleotide excision repair." Nucleic Acids Research 52.2 (2024): 677-689. https://doi.org/10.1093/nar/gkad1104
The article indicates that XPA is a core factor of NER, but it is absent in organisms such as Arabidopsis thaliana. Studies have shown that humans, fruit flies and worms can still perform inefficient excision and repair in the absence of XPA, revealing the special position of XPA in evolution and its non-absolute necessity in repair.
Creative Biolabs: XPA Antibodies for Research
Creative Biolabs specializes in the production of high-quality XPA antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom XPA Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our XPA antibodies, custom preparations, or technical support, contact us at email.
Reference
- Krasikova, Yuliya S., Olga I. Lavrik, and Nadejda I. Rechkunova. "The XPA Protein—Life under Precise Control." Cells 11.23 (2022): 3723. https://doi.org/10.3390/cells11233723
Anti-XPA antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-CCT6A/B Recombinant Antibody (CBXC-0168) (CBMAB-C5570-CQ)

-
Rabbit Anti-ALDOA Recombinant Antibody (D73H4) (CBMAB-A2314-YC)

-
Mouse Anti-ALB Recombinant Antibody (V2-180650) (CBMAB-A2186-YC)

-
Mouse Anti-ENO2 Recombinant Antibody (H14) (CBMAB-E1341-FY)

-
Mouse Anti-AKT1 Recombinant Antibody (V2-180546) (CBMAB-A2070-YC)

-
Mouse Anti-CD24 Recombinant Antibody (SN3) (CBMAB-C1037-CQ)

-
Mouse Anti-CFL1 (Phospho-Ser3) Recombinant Antibody (CBFYC-1770) (CBMAB-C1832-FY)

-
Mouse Anti-DISP2 Monoclonal Antibody (F66A4B1) (CBMAB-1112CQ)

-
Rat Anti-CD63 Recombinant Antibody (7G4.2E8) (CBMAB-C8725-LY)

-
Rabbit Anti-BRCA2 Recombinant Antibody (D9S6V) (CBMAB-CP0017-LY)

-
Rabbit Anti-ABL1 (Phosphorylated Y185) Recombinant Antibody (V2-443434) (PTM-CBMAB-0001YC)

-
Mouse Anti-ATP1B1 Recombinant Antibody (E4) (CBMAB-0463-LY)

-
Rabbit Anti-DLK1 Recombinant Antibody (9D8) (CBMAB-D1061-YC)

-
Mouse Anti-DES Monoclonal Antibody (440) (CBMAB-AP1857LY)

-
Rat Anti-EPO Recombinant Antibody (16) (CBMAB-E1578-FY)

-
Mouse Anti-AZGP1 Recombinant Antibody (CBWJZ-007) (CBMAB-Z0012-WJ)

-
Mouse Anti-CRTAM Recombinant Antibody (CBFYC-2235) (CBMAB-C2305-FY)

-
Rabbit Anti-AKT2 (Phosphorylated S474) Recombinant Antibody (V2-556130) (PTM-CBMAB-0605LY)

-
Mouse Anti-CASQ1 Recombinant Antibody (CBFYC-0863) (CBMAB-C0918-FY)

-
Mouse Anti-AAV-5 Recombinant Antibody (V2-503416) (CBMAB-V208-1402-FY)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot








