Coronavirus
Creative Biolabs is an award-winning antibody-related products and services supplier, we offer a comprehensive antibody production and service portfolio for the coronavirus research and diagnosis. Our antibody development services include from antigen design and production, custom antibody generation to antibody analysis, our expert scientists and talented technical support team have got you covered all the way.
Introduction of Coronavirus
Coronaviruses are a group of viruses that can cause severe diseases in mammals and birds. In humans, it often causes diseases from mild respiratory tract infections to lethal death. Symptoms in other species are different. Coronaviruses can cause an upper respiratory tract disease in chickens, while in cows and pigs they cause diarrhea. To date, there is no effective vaccine or antiviral drug to prevent or treat human coronavirus infections.
Coronaviruses belong to the Orthocoronavirinae subfamily of the Coronaviridae family, Nidovirales order, and Riboviria realm. They have a nucleocapsid of helical symmetry and a positive-sense single-stranded RNA genome that is the largest known RNA viruses and the genome size ranges from approximately 27 to 34 kilobases. Coronavirus is covered by club-shaped protein spikes.
The earliest human coronaviruses were first discovered in the late 1960s ones from chickens infected with an infectious bronchitis virus and human patients with the common cold. Subsequently, other members of this family have been found, including SARS-CoV, HCoV NL63, HKU1, MERS-CoV, and SARS-CoV-2 (also known as 2019-nCoV). Most of theses coronaviruses have caused serious respiratory tract infections.
Coronaviruses are large pleomorphic spherical particles with bulbous surface projections. The diameter of the virus particles is approximately 120 nm. The envelope of the virus is composed of a lipid bilayer where the membrane (M), envelope (E) and spike (S) structural proteins are anchored, which in electron micrographs shows a distinct pair of electron dense shells. A subset of coronaviruses also presents a shorter spike-like surface protein called hemagglutinin esterase (HE). The nucleocapsid is located in the envelope consisting of multiple copies of the nucleocapsid (N) protein that are bound to the positive-sense single-stranded RNA genome in a continuous beads-on-a-string type conformation. The lipid bilayer envelope, membrane proteins, and nucleocapsid play an important role in protecting the virus when it is outside the host cell.
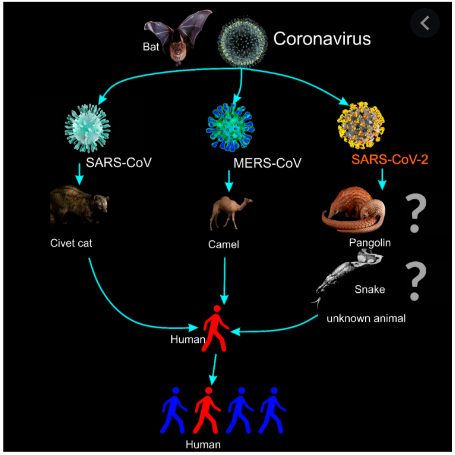 Fig.1 The illustration for the transmission of coronaviruses and the 2019 novel Coronavirus (2019-nCoV or SARS-CoV-2).1,3
Fig.1 The illustration for the transmission of coronaviruses and the 2019 novel Coronavirus (2019-nCoV or SARS-CoV-2).1,3
Human Coronaviruses
Different human coronaviruses present different risks. Some fatal types such as the MERS-CoV can kill more than 30% of those infected, and some are relatively harmless, such as the HCoV-OC43 only resulting in common cold. Major symptoms infected by Coronaviruses include fever, sore throat from swollen adenoids, pneumonia and bronchitis. Particularly, the SARS-CoV coronavirus discovered in 2003 has a unique pathogenesis (upper and lower respiratory tract infections) that can lead to severe acute respiratory syndrome (SARS). Among seven known strains of human coronaviruses, four cause the mild symptoms of the common cold, including human coronavirus OC43 (HCoV-OC43), human coronavirus HKU1, human coronavirus NL63 (HCoV-NL63, New Haven coronavirus) and human coronavirus 229E (HCoV-229E). Other three strains can cause severe symptoms including severe acute respiratory syndrome coronavirus (SARS-CoV or "SARS-classic", broke out in 2003), Middle East respiratory syndrome-related coronavirus (MERS-CoV, broke out in 2012, 2015 and 2018), and the latest severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, 2019-nCoV or "novel coronavirus 2019", broke out in 2019).
Latest SARS-CoV-2 (2019-nCov)
SARS-CoV-2 (also known as 2019 novel coronavirus, 2019-nCoV) is the seventh coronavirus known to infect humans, which is a positive-sense single-stranded RNA virus. So far, it has been designated a Public Health Emergency of International Concern by the World Health Organization (WHO) due to it is contagious in humans and has caused the pandemic infection of coronavirus disease.
- Genetic Characteristics and Virion Structure
SARS-CoV-2 belongs to the order Nidovirales, family Coronaviridae, and the subfamily Coronavirinae. The whole-genome sequencing and the genetic analysis showed that SARS-CoV-2 is genetically related to SARS-CoV of the 2003 outbreak. It was also found to be closely related to the genus Betacoronavirus and was a distinct clade in lineage B of the subgenus Sarbecovirus, collectively with two other bat-derived SARS-CoV-like strains. To date, the origin of the virus is not understood clearly. The latest research found that a membrane exopeptidase called angiotensin-converting enzyme 2 (ACE 2), is the receptor used by SARS-CoV-2 to enter into the human cells, much like its predecessor (SARS-CoV).
Studies found that SARS-CoV-2 was closely related (with 88% similarity) to two bat-origin SARS-like CoVs (Bat-SL-CoV), Bat-SL-CoVZC45 and Bat-SL-CoVZXC21. Importantly, SARS-CoV-2 was found to be non-identical with SARS-CoV (about 79% similarity) and MERS-CoV (about 50% similarity).
SARS-CoV-2 has four structural proteins including the S (spike), E (envelope), M (membrane), N (nucleocapsid) proteins and hemagglutinin esterase (HE) (only found in some beta-CoVs). The nucleocapsid protein complexes with the genome RNA to form a helical capsid structure in the viral envelope. Trimers of the spike proteins form the peplomers embedded in the envelope, endowing the virion its corona or crown-like morphology. In some CoV virions, the HE protein can form smaller spikes at the membrane. The “M” and “E” are a type of transmembrane proteins involved in virus assembly.
CoV virions are spherical and approximately 50-200 nanometres in diameter. The typical characteristic features of CoVs are the club-shaped spike projections emanating from the surface of the virion. These spikes are a prominent feature of virions that make them look like the corona, hence the name coronaviruses. CoVs have helically symmetrical nucleocapsids located in the envelope of the virion, which are unusual among positive-sense RNA viruses, however, a way more common for negative-sense RNA viruses.
- Epidemiology
To date, seven CoV species can infect human beings, and only MERS-CoV and SARS-CoV have been able to cause severe human disease. They may cause serious disease in immunocompromised individuals. At present, the SARS-CoV-2 (nCoV-19) infection is life-threatening, and can cause deaths. They possess wide host adaptability and can cause severe infections in humans, birds, livestock, masked palm civets, mice, dogs, cats, camels, pigs, chickens, and bats, wherein they typically cause respiratory and gastrointestinal sickness. Four human CoVs (HCoVs) (HCoV 229E, NL63, OC43, and HKU1) are endemic globally and account for 10% to 30% of upper respiratory tract infections in adults. Ecologically, there are several types of CoVs, and most of them were discovered in bats, suggesting that bats may be natural reservoirs for a lot of these viruses. Peri-domestic mammals can also serve as intermediate hosts to facilitate virus recombination, mutation, and genetic variations. SARS-CoV-2 presently is fast spreading to different countries throughout the world including European, American, Asian, and some African countries.
- Transmission
So far, the exact mechanisms of transmission are uncertain, human-to-human transmission can arise, and the risk of airborne spread appears imminent. Furthermore, SARS-CoV-2 may be transmitted from bats to palm civets or dromedary camels and thereby spill over into human beings. The infection pathways may include reintroduction into human beings from an animal reservoir, persistent infection in previously sick people, or the laboratory strains.
Generally, SARS-CoV-2 can transmit through direct or indirect contact of mucous membranes (such as the eyes, nose, or mouth, etc) with infectious respiratory droplets or fomites. Transmission risks increase with period and proximity with the infected patients. The survival time of SARS-CoV-2 within the environment is unknown currently. However, recent studies revealed that SARS-CoV can live up to two weeks after drying and five days at temperatures of 22-25°C and 40-50% relative humidity, with a gradual decrease in the viability of the virus. The viability of SARS-CoV will decline significantly after 24 hours at 38°C and 80-90% relative humidity. It may remain alive on distinct surfaces for 48 hours at 20°C and 40% relative humidity, despite viability reduced to 8 hours at 30°C and 80% relative humidity conditions. All above evidence showed that under a low temperature, low humidity conditions can facilitate the virus survival in the environment.
SARS-CoV-2 can be transmitted rapidly among humans and exhibits high potential for a pandemic according to the evidence of an increasing incidence of infections and the possibility of transmission by asymptomatic carriers. In addition, the advancement and convenience of global travel also promote the worldwide spread of SARS-CoV-2.
- Pathogenesis
SARS-CoV-2 demonstrates versatile host ranges and tissue tropism. The attachment of the virus to the host cell is initiated by interactions between the “S” protein and its receptor. The sites of the receptor-binding domain (RBD) in the S1 vicinity of a CoV’s “S” protein are different among the virus strains. Some have the RBD at the N-terminus of S1 (MHV) while others have the RBD at the C-terminus of S1.
Structural, enzymatic, and accent proteins of SARS-CoV-2 play critical roles in the pathogenesis of the CoV disease (COVID). Structural proteins functions in the maintenance of virion shape and morphogenesis and involve in the viral spread in vivo and antagonizing host cellular and immune responses. Small accent proteins have an enzymatic role in RNA metabolism or protein processing, which further favors the viruses in antagonizing host immune responses during infections.
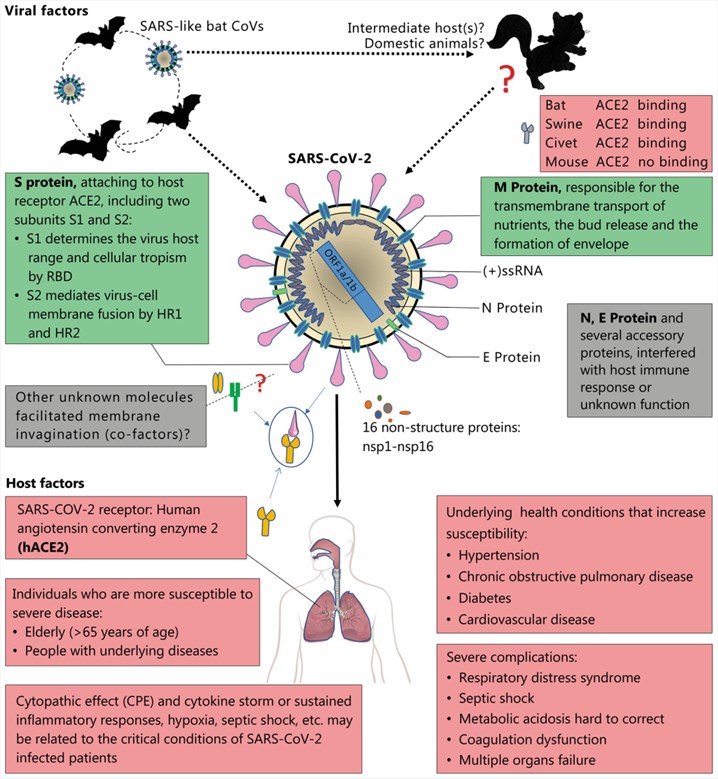 Fig.2 Viral and host factors that influence the pathogenesis of SARS-CoV-2.2,3
Fig.2 Viral and host factors that influence the pathogenesis of SARS-CoV-2.2,3
- Clinical Signs
The incubation period of COVID may range from one to two weeks. The clinical manifestations in patients infected with SARS-CoV-2 include mild, moderate, severe and rapidly progressive and fulminant disease. The most common symptoms include fever, cough, myalgia or fatigue, and atypical symptoms include sputum, headache, hemoptysis and diarrhea. About half of the patients present dyspnea and all patients have pneumonia. Serious complications include heart failure, respiratory failure, and liver failure in elderly patients.
Respiratory failure is the most severe problem of COVID, and at least half of the patients require supplemental oxygen during the intense phase. Additionally, about 20% of patients progress to acute respiratory distress syndrome need invasive mechanical ventilator support. Deaths commonly occur as early as day 4 and as overdue as 108 days once the onset of symptoms. Research showed that the virus shedding from the respiratory tract was around day 10 and later declined. Immunoglobulin G (IgG) antibodies can be detected at 10-15 days after the onset of symptoms. The severity of the disease was significantly associated with increasing age, with increased mortality (up to 50%) in patients over 60 years of age.
- Laboratory Diagnosis
Various human clinical specimens (such as the sputum, throat swab, nasal secretions, feces, blood/serum/plasma) can be used for the diagnosis of SARS-CoV-2 in Laboratory tests. Diagnosis methods include real-time polymerase chain reaction (RT-PCR), viral cultural techniques for the isolation of virus from clinical specimens, immunological tests for the detection of antibodies and enzyme-linked immunosorbent assay (ELISA), indirect fluorescent antibody technique, rapid immunochromatographic tests, and immunofluorescence techniques. Other tests such as the flow-cytometry analysis for CD4+ and CD8+ T cell counts, chest radiography (pneumonia), complete blood picture (to demonstrate lymphopenia), and serum biochemistry (serum protein and others) can be used as assists in the diagnosis.
- Treatment
Currently, there is no single specific anti-viral therapy available against COVID. The cases of SARS-CoV-2 infection are continuously increasing throughout the world since its outbreak in China. M protein of SARS-CoV-2 presently has been used as a potential target that may be inhibited by the already available and approved drugs. Studies have displayed that the available drugs against SARS-CoV-2 “P” and “L” proteins have the potential to inhibit the catalytic area and inactivate the virus.
Anti-viral treatment with interferon-alpha inhalation, anti-retroviral drugs, and arbidol is recommended. Treated with a corticosteroid and gamma globulin for three to five days can rest respiratory rate greater than 30 per minute, or make oxygen saturation was under 93% without oxygen. Probiotics are used to relieve gastrointestinal illness. Quinolones and higher generation cephalosporins (oral and intravenous) can be used if fever lasted for more than seven days or C-reactive protein levels were 30 mg/L or more (normal range: 0-8 mg/L). After the course of treatment, the patients can be discharged from the hospital only after two negative results of the samples collected with 24-hour interval using an RT-PCR. Other drugs such as the chloroquine, hydroxychloroquine, azithromycin, and favipiravir are under clinical trials to treat serious infections caused by SARS-CoV-2. In addition, protease inhibitors, RNA interference, monoclonal antibody, synthetic peptides, and vaccines are also being developed.
- Control and Preventive Measures
To control the spread of respiratory secretions, all people with signs and symptoms of respiratory infection, irrespective of presumed cause, have to be advised to cover the nose and mouth while coughing or sneezing. People must be advised to use tissues to contain respiratory secretions and avoid them within the nearest waste disposal container.
Because of the virus can survive several days in the environment, the premises and areas contaminated with SARS-CoV-2 have to be wiped clean earlier than their re-use with disinfectants containing antimicrobial agents. Many antimicrobial agents have been confirmed against different CoVs, such as the sodium hypochlorite and ethanol.
There are three main prevention strategies for SARS. Firstly, stop the transmission of infection to increase personal protection for individuals. Secondly, detect the SARS infection as early as possible and referring suspected individuals to the quarantine or emergency room in the nearby medical center if needed. Finally, focus on restoration and rehabilitation. Early recognition of the disease, rapid diagnosis, isolation, and stringent infection control measures are the keys to control disease spread.
What Can We Do For You?
At Creative Biolabs, we prioritize your scientific success and are here to help, if you are struggling to find a suitable antibody for your coronaviruses research and diagnosis. Creative Biolabs is a leader in the field of antibody development, and our goal is to provide you with the most affordable and high-quality antibody products and antibody-related services to ensure your satisfaction. We serve the diverse needs of our clients and we can tailor specific service packages to fit your timeline and R&D budget. Please contact us for more information and a detailed quote.
We offer the following products for coronavirus detection and diagnosis:
References
- Nguyen, Trieu, Dang Duong Bang, and Anders Wolff. "2019 novel coronavirus disease (COVID-19): paving the road for rapid detection and point-of-care diagnostics." Micromachines 11.3 (2020): 306.
- Guo, Yan-Rong, et al. "The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status." Military medical research 7 (2020): 1-10.
- Distributed under Open Access license CC BY 4.0, without modification.


