Cell Cycle Staining Flow Cytometry
Cell Cycle Introduction
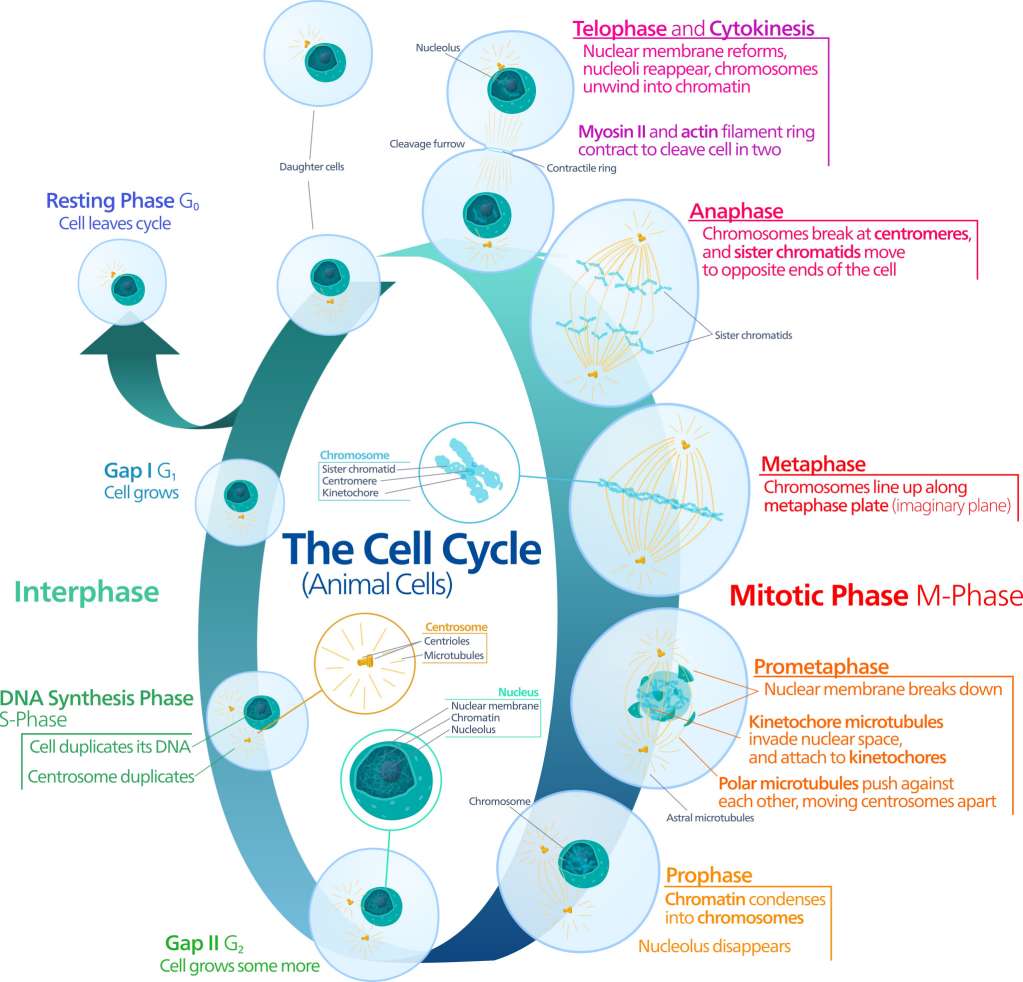
Cell cycle includes two distinct and specific phases, interphase, consisting of G1, S, and G2, and mitotic phase, M. During cell cycle progression, proliferating cells sequentially undergo a transition of G1→S→G2→M phases. The cell grows in G1 phase, accumulates the energy necessary for duplication and replicates cellular DNA in S phase, and prepares to divide in G2 phase. Each phase of the cell cycle is tightly regulated.
There are two checkpoints to detect potential DNA damage and allow it to be repaired before cell divides. If the damage cannot be repaired, a cell becomes targeted for apoptosis. Cells can also reversibly stop dividing and temporarily enter G0 phase. The first checkpoint is at the end of G1, making the decision whether a cell should enter S phase and divide, delay division, or enter G0. The second checkpoint, at the end of G2, triggers mitosis whether a cell has all the necessary components.
Cell Cycle Analysis by Flow cytometry
Assessing cell cycle distribution and cell proliferation is extremely important for researchers to study cell growth differentiation, senescence and apoptosis. The most commonly used technique to achieve these goals is flow cytometry. Through this assay, a fluorescent dye that binds to DNA is incubated with a single cell suspension of permeabilized or fixed cells. Since the dye binds to DNA stoichiometrically, the amount of fluorescent signal is directly proportional to the amount of DNA. Because of the alterations that occur during the cell cycle, analysis of DNA content allows discrimination between G1, S, G2 and M phases. The DNA of yeast, plant, bacterial cells, and mammalian can be stained by a variety of DNA binding dyes such as propidium iodide (PI), 7-amino actinomycin-D (7-AAD), Hoechst 33342 and 33258, and 4’6’-diamidino-2-phenylindole (DAPI), BrdU, TO-PRO-3 Iodide and so forth.

Analysis Methods
Cell proliferation can be measured by flow cytometry using different methods. However, it is important to recognize that simple single stained flow cytometry analysis using 7-AAD or PI is unable to distinguish between cells in G1 or G2 from those in very early or very late S phase, and similarly those in G2 or M. Therefore, it is sometimes necessary to combine these dyes with a proliferative marker. One method is to stain with an antibody against a proliferation marker such as Ki67. Alternatively, you can incubate your cells with BrdU, which incorporates into DNA during S-phase of the cell cycle. Incorporated BrdU can be detected using fluorescently labeled anti-BrdU antibodies. When combined with a DNA stain such as PI or DAPI, the relative proportion of cells in S-phase can be determined.
In summary, there are four commonly used methods to analyze the cell cycle by flow cytometry.
- The first two are based on univariate analysis of cellular DNA content following cell staining with either PI or DAPI and deconvolution of the cellular DNA content frequency histograms. This approach reveals distribution of cells in three major phases of the cycle (G1 vs S vs G2/M) and makes it possible to detect apoptotic cells with fractional DNA content.
- The third approach is based on the bivariate analysis of DNA content and proliferation-associated proteins.
- The fourth procedure relies on the detection of BrdU incorporation to label the DNA-replicating cells.
The method used will depend on the experiment and the information required.
Classic Workflow
- Harvest cells and wash with PBS
- Cell fixation or permeabilization
- Wash with PBS
- Treat with RNase
- Treat with PI/BrdU
- Flow cytometry analysis
Protocol of Cell Cycle Staining Flow Cytometry is available for you. It mainly includes the following parts:
- Protocol of Immunofluorescence staining of cells in combination with propidium iodide staining of cells for cell cycle analysis
- Protocol of propidium iodide staining of cells for cell cycle analysis
- Protocol of BrdU staining of cells for cell cycle analysis
If you get in trouble, please refer to our Troubleshooting of Cell Cycle Staining Flow Cytometry.


