MGAM Antibodies
Background
MGAM (Maltase-Glucoamylase) is a bifunctional glycoside hydrolase located in the brush border membrane of the human small intestine, existing in the form of a dimer. This enzyme plays a key role in the final stage of carbohydrate digestion by hydrolyzing starchy substrates to produce glucose through the synergistic action of its N-terminal and C-terminal catalytic domains. Due to its significant role in blood glucose regulation, MGAM has become one of the important targets for the development of type 2 diabetes drugs. After its gene sequencing was completed in 2000, researchers analyzed the three-dimensional structure of the MGAM and inhibitor complex through X-ray crystallography. This not only revealed the specific substrate recognition mechanism of the enzyme but also provided an accurate molecular blueprint for the design of targeted drugs. The continuous research on the catalytic mechanism and allosteric regulation of this enzyme has significantly advanced our understanding of the regulatory mechanisms of digestive physiology and enzyme kinetics.
Structure of MGAM
MGAM is a large transmembrane glycoprotein with a molecular weight of approximately 210-230 kDa. Its precise molecular weight may vary depending on different transcripts and the degree of glycosylation modification.
| Species | Human | Mouse | Rat |
| Molecular Weight (kDa) | 210-230 | 208-225 | 209-227 |
| Primary Structural Differences | Contains two catalytic Nt - MGAM/Ct - MGAM structure domain | Sequence is highly conserved structure domain | Catalytic site highly homologous with humans |
This enzyme is composed of 1,857 amino acid residues and adopts a typical (β/α) 8-barrel-shaped folding structure. Its N-terminal and C-terminal each contain an independent catalytic domain, which are connected in series through a flexible linking peptide to form a synergistic active center. The key acidic residues in the structure (such as D542 and D2039) directly participate in the hydrolysis of glycosidic bonds, while the aromatic amino acids in the substrate-binding channels precisely locate starch molecules through stacking. This multi-domain assembly mode enables it to efficiently hydrolyze starch substrates of different chain lengths.
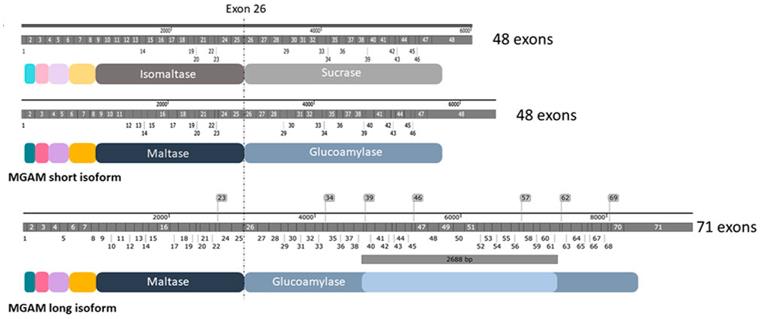 Fig. 1 Structural features and protein characterization of SI and MGAM.1
Fig. 1 Structural features and protein characterization of SI and MGAM.1
Key structural properties of MGAM:
- Bifunctional enzyme structure consisting of two catalytic domains, N-terminal and C-terminal
- Each domain adopts the classical glycoside hydrolase configuration of (β/α)₈barrel folding
- Active center contains highly conserved catalytic triad (acidic residues)
- The substrate combination channel by aromatic amino acid starch molecular precision positioning
Functions of MGAM
The core function of the MGAM gene-encoded protein is to catalyze the hydrolysis of starch into glucose. Its main physiological functions include:
| Function | Description |
| Starch digestion | Hydrolysis of α-1, 4-glycosidic bonds at the brush border of the small intestine breaks down the oligosaccharide into absorbable glucose. |
| Blood glucose regulation | By controlling the final digestion rate of carbohydrates, it directly affects the postprandial blood sugar level. |
| Dual-function synergy | The N-terminal domain prefers to hydrolyze short-chain maltose, while the C-terminal domain is more effective in hydrolyzing long-chain starch. |
| Metabolic regulation | As a therapeutic target for diabetes, the inhibition of its activity can delay glucose absorption. |
| Nutrient absorption | Complete the final step of carbohydrate digestion and provide the main energy source for the body. |
The catalytic efficiency of this enzyme shows a typical substrate chain length dependence. Its dual domains achieve comprehensive hydrolysis of starchy substances through differential substrate specificity. This division of labor and cooperation mechanism significantly improves the energy uptake efficiency of the digestive system.
Applications of MGAM and MGAM Antibody in Literature
1. Chasib Mezher, Rawaa, et al. "Pan‐cancer analysis and oncogenic implications of MGAM and MGAM2: Toward precision oncology and drug repurposing in colorectal cancer." Journal of Cell Communication and Signaling 19.3 (2025): e70042. https://doi.org/10.1002/ccs3.70042
Research has found that MGAM and its homologous genes are abnormally expressed in various cancers and are related to key pathways such as metabolism and immunity, which can serve as potential biomarkers and therapeutic targets.
2. Vincent-Chong, Vui King, et al. "Genome wide analysis of chromosomal alterations in oral squamous cell carcinomas revealed over expression of MGAM and ADAM9." PloS one 8.2 (2013): e54705. https://doi.org/10.1371/journal.pone.0054705
This study, for the first time, analyzed the genome of oral squamous cell carcinoma through high-throughput microarray technology and discovered multiple frequently amplified regions. The genes MGAM and ADAM9 were significantly amplified and overexpressed in tumors, especially the expression of MGAM increased by 6.6 times, which is expected to become a new biomarker.
3. Sathe, Gajanan, et al. "Urinary glycoproteomic profiling of non-muscle invasive and muscle invasive bladder carcinoma patients reveals distinct N-glycosylation pattern of CD44, MGAM, and GINM1." Oncotarget 11.34 (2020): 3244. https://doi.org/10.18632/oncotarget.27696
This study, through urine glycoproteomics analysis, found that the N-glycosylation patterns of proteins such as CD44 and MGAM in bladder cancer show specific changes at different pathological stages, providing new insights into the diagnosis and disease progression mechanism of bladder cancer.
4. Wang, Zheng, Guancheng Quan, and Gang He. "Edge-Oriented Compressed Video Super-Resolution." Sensors 24.1 (2023): 170. https://doi.org/10.3390/s24010170
This article develops a highly sensitive fluorescence-based biosensor using quantum dots conjugated with plastic antibodies to detect myoglobin at femtomolar concentrations, providing a cost-effective, selective, and stable alternative for early myocardial infarction diagnosis in human serum.
5. Tannous, Stephanie, et al. "Interaction between the α-glucosidases, sucrase-isomaltase and maltase-glucoamylase, in human intestinal brush border membranes and its potential impact on disaccharide digestion." Frontiers in molecular biosciences 10 (2023): 1160860. https://doi.org/10.3389/fmolb.2023.1160860
This study confirmed that intestinal α -glycosidase SI and MGAM interact in the brush margin membrane to form a heterocomplex, jointly regulating the final step of starch digestion, among which SI plays a major role.
Creative Biolabs: MGAM Antibodies for Research
Creative Biolabs specializes in the production of high-quality MGAM antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom MGAM Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our MGAM antibodies, custom preparations, or technical support, contact us at email.
Reference
- Tannous, Stephanie, et al. "Interaction between the α-glucosidases, sucrase-isomaltase and maltase-glucoamylase, in human intestinal brush border membranes and its potential impact on disaccharide digestion." Frontiers in molecular biosciences 10 (2023): 1160860. https://doi.org/10.3389/fmolb.2023.1160860
Anti-MGAM antibodies
 Loading...
Loading...
Hot products 
-
Rat Anti-CD300A Recombinant Antibody (172224) (CBMAB-C0423-LY)

-
Mouse Anti-CD24 Recombinant Antibody (2Q1282) (CBMAB-C1624-CN)

-
Mouse Anti-ASB9 Recombinant Antibody (1D8) (CBMAB-A0529-LY)

-
Mouse Anti-ARHGDIA Recombinant Antibody (CBCNA-009) (CBMAB-R0415-CN)

-
Rabbit Anti-AKT2 (Phosphorylated S474) Recombinant Antibody (V2-556130) (PTM-CBMAB-0605LY)

-
Mouse Anti-CD247 Recombinant Antibody (6B10.2) (CBMAB-C1583-YY)

-
Mouse Anti-BrdU Recombinant Antibody (IIB5) (CBMAB-1038CQ)

-
Mouse Anti-CCS Recombinant Antibody (CBFYC-1093) (CBMAB-C1150-FY)

-
Mouse Anti-ALB Recombinant Antibody (V2-55272) (CBMAB-H0819-FY)

-
Mouse Anti-AAV-5 Recombinant Antibody (V2-503417) (CBMAB-V208-1369-FY)

-
Mouse Anti-ABIN2 Recombinant Antibody (V2-179106) (CBMAB-A0349-YC)

-
Mouse Anti-ADAM29 Recombinant Antibody (V2-179787) (CBMAB-A1149-YC)

-
Mouse Anti-CCT6A/B Recombinant Antibody (CBXC-0168) (CBMAB-C5570-CQ)

-
Rabbit Anti-BRCA2 Recombinant Antibody (D9S6V) (CBMAB-CP0017-LY)

-
Mouse Anti-8-oxoguanine Recombinant Antibody (V2-7719) (CBMAB-1898CQ)

-
Mouse Anti-ATP5F1A Recombinant Antibody (51) (CBMAB-A4043-YC)

-
Mouse Anti-CCDC6 Recombinant Antibody (CBXC-0106) (CBMAB-C5397-CQ)

-
Mouse Anti-ACLY Recombinant Antibody (V2-179314) (CBMAB-A0610-YC)

-
Mouse Anti-8-oxoguanine Recombinant Antibody (V2-7697) (CBMAB-1869CQ)

-
Mouse Anti-ABL2 Recombinant Antibody (V2-179121) (CBMAB-A0364-YC)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






