CENPA Antibodies
Background
CENPA (centromerin A) is a histone H3 variant that mainly exists in the centromeric region of eukaryotes and constitutes an important structural basis of chromatin. This protein forms a special nucleosome by replacing the conventional histone H3, which not only maintains the stability of the chromatin of the centromere but also provides a binding platform for centromere assembly proteins, thereby ensuring the precise separation of chromosomes during cell division. As the core carrier of centromeric epigenetic markers, CENPA mediates centromeric localization through its histone folding domain, while its amino-terminal tail participates in regulating protein interactions. In 1998, researchers first identified CENPA in mammals. Its unique centromer-specific assembly mechanism not only deepened our understanding of epigenetic regulation but also provided a key molecular basis for the study of chromosomal abnormality diseases.
Structure of CENPA
CENPA is a key histone variant with a molecular weight of approximately 15-16 kDa. The molecular weight of this protein varies slightly among different species, mainly due to the diversity of its N-terminal tail sequence.
| Species | Human | Mouse | African clawed toad | Fruit fly | Yeast |
| Molecular Weight (kDa) | 15.8 | 15.9 | 16.1 | 16.5 | 16.3 |
| Primary Structural Differences | Typical CATD domains | Highly homologous to humans | Conservative core area | The CID domain is specific | The most simplified variant form |
This protein is composed of approximately 140 amino acids, and its core feature lies in the histone folding domain, which forms a unique dimerization interface. The core region of CENPA contains a specific Loop1 segment and antiparallel α -helical bundles, which can preferentially assemble with histone H4 to form heterodimers. Its N-terminal tail is shorter and the sequence is more variable than that of the conventional H3. This characteristic affects the assembly of the advanced structure of the chromatin in the silk grains and the maintenance of the epigenetic state.
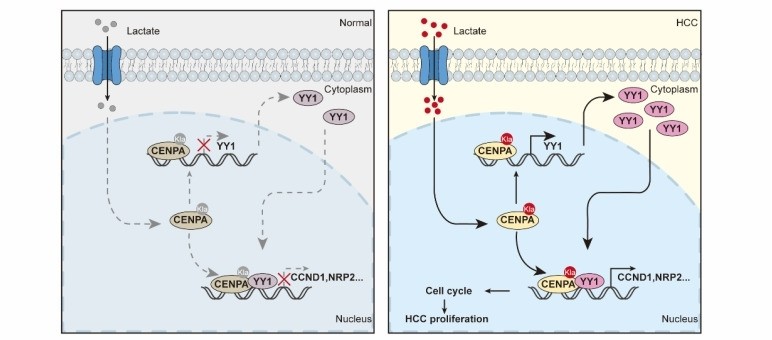 Fig. 1 CENPA Drives HCC Progression through Transcriptional Activation.1
Fig. 1 CENPA Drives HCC Progression through Transcriptional Activation.1
Key structural properties of CENPA :
- Characteristic histone folding domains
- Specific Loop 1 area constitute two polymerization interface
- Histone chaperone recognition domain (CATD) is used for localization
Functions of CENPA
The core function of CENPA is to guide the localization of filaments during cell division and ensure the correct separation of chromosomes. In addition, it is also involved in regulating chromatin stability and epigenetic memory.
| Function | Description |
| Centromere identity is determined | By replacing the conventional histone H3, specific locations of centromeres are marked, providing an epigenetic basis for kinetosomal assembly. |
| Precise separation of chromosomes | Centromer-specific nucleosomes are formed to ensure that chromatids are evenly pulled towards the two poles by the spindle fibers during cell mitosis and meiosis. |
| Maintain genomic stability | Its normal function can prevent chromosomal misseparation and aneuploidy, thereby inhibiting the occurrence of genomic diseases such as tumors. |
| Epigenetic information carrying | Passing centromere characteristics to progeny cells without relying on DNA sequence changes is a typical epigenetic regulatory mechanism. |
| Regulate the higher-order structure of chromatin | Acting synergistically with other centromere proteins to help the centromere region form a tight chromatin structure to perform its function. |
The localization of CENPA at centromeres is not dependent on DNA sequences but is dominated by epigenetic mechanisms, making it a key determinant for maintaining chromosomal genetic stability.
Applications of CENPA and CENPA Antibody in Literature
1. Liao, Jingyu, et al. "CENPA functions as a transcriptional regulator to promote hepatocellular carcinoma progression via cooperating with YY1." International journal of biological sciences 19.16 (2023): 5218. https://doi.org/10.7150/ijbs.85656
This study reveals that CENPA is highly expressed in hepatocellular carcinoma (HCC) and is associated with a poor prognosis. CENPA is activated through lactation modification at the K124 site and collaborates with YY1 to transcriptionally regulate the expression of CCND1 and NRP2, thereby promoting the progression of HCC. Targeting the CENPA-YY1-CCND1/NRP2 axis may be a potential therapeutic strategy.
2. Yu, Qi-Ying, et al. "CENPA regulates tumor stemness in lung adenocarcinoma." Aging (Albany NY) 14.13 (2022): 5537. https://doi.org/10.18632/aging.204167
This study constructed a risk model of lung adenocarcinoma through bioinformatics analysis and identified CENPA. This gene is not only a key hub gene, but also serves as a tumor stem regulatory factor and prognostic marker, playing a significant role in the development of lung adenocarcinoma.
3. Li, Junwu, et al. "High CENPA expression in papillary renal cell carcinoma tissues is associated with poor prognosis." BMC urology 22.1 (2022): 157. https://doi.org/10.1186/s12894-022-01106-4
This study confirmed that CENPA is significantly highly expressed in renal papillary cell carcinoma (PRCC) and is closely related to advanced clinical stage and poor prognosis. Analysis shows that CENPA is an independent prognostic factor and can serve as a potential prognostic molecular marker and therapeutic target for PRCC.
4. Wang, Bo, et al. "CENPA acts as a prognostic factor that relates to immune infiltrates in gliomas." Frontiers in Neurology 13 (2022): 1015221. https://doi.org/10.3389/fneur.2022.1015221
This study confirmed that CENPA is abnormally highly expressed in glioma and is associated with a poor prognosis. The expression level of CENPA is closely related to clinical features such as tumor grade and IDH mutation. Its overexpression can promote the proliferation and migration of tumor cells and affect the tumor immune microenvironment, and can be used as a potential independent prognostic marker and therapeutic target.
5. Wang, Qi, et al. "CENPA promotes clear cell renal cell carcinoma progression and metastasis via Wnt/β-catenin signaling pathway." Journal of translational medicine 19.1 (2021): 417. https://doi.org/10.1186/s12967-021-03087-8
This study confirmed that in clear cell renal cell carcinoma (ccRCC), CENPA, as a representative of the CENP family, is significantly highly expressed and is associated with clinicopathological grade and prognosis. Studies have shown that CENPA promotes the proliferation and metastasis of ccRCC by accelerating the cell cycle and activating the Wnt/β-catenin signaling pathway, suggesting that it can serve as a potential therapeutic target.
Creative Biolabs: CENPA Antibodies for Research
Creative Biolabs specializes in the production of high-quality CENPA antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom CENPA Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our CENPA antibodies, custom preparations, or technical support, contact us at email.
Reference
- Liao, Jingyu, et al. "CENPA functions as a transcriptional regulator to promote hepatocellular carcinoma progression via cooperating with YY1." International journal of biological sciences 19.16 (2023): 5218. https://doi.org/10.7150/ijbs.85656
Anti-CENPA antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-CRYAB Recombinant Antibody (A4345) (CBMAB-A4345-YC)

-
Mouse Anti-BAD (Phospho-Ser136) Recombinant Antibody (CBYY-0138) (CBMAB-0139-YY)

-
Mouse Anti-AKT1 Recombinant Antibody (V2-180546) (CBMAB-A2070-YC)

-
Rabbit Anti-ADRA1A Recombinant Antibody (V2-12532) (CBMAB-1022-CN)

-
Mouse Anti-DMPK Recombinant Antibody (CBYCD-324) (CBMAB-D1200-YC)

-
Mouse Anti-APCS Recombinant Antibody (CBYC-A663) (CBMAB-A3054-YC)

-
Mouse Anti-CD33 Recombinant Antibody (6C5/2) (CBMAB-C8126-LY)

-
Mouse Anti-CCNH Recombinant Antibody (CBFYC-1054) (CBMAB-C1111-FY)

-
Rabbit Anti-CAMK2A Recombinant Antibody (BA0032) (CBMAB-0137CQ)

-
Mouse Anti-ADGRE5 Recombinant Antibody (V2-360335) (CBMAB-C2088-CQ)

-
Mouse Anti-BAX Recombinant Antibody (CBYY-0216) (CBMAB-0217-YY)

-
Mouse Anti-CIITA Recombinant Antibody (CBLC160-LY) (CBMAB-C10987-LY)

-
Mouse Anti-ENO1 Recombinant Antibody (8G8) (CBMAB-E1329-FY)

-
Mouse Anti-CD24 Recombinant Antibody (HIS50) (CBMAB-C10123-LY)

-
Mouse Anti-dsDNA Recombinant Antibody (22) (CBMAB-AP1954LY)

-
Mouse Anti-BRD3 Recombinant Antibody (CBYY-0801) (CBMAB-0804-YY)

-
Human Anti-SARS-CoV-2 S1 Monoclonal Antibody (CBFYR-0120) (CBMAB-R0120-FY)

-
Rabbit Anti-BRCA2 Recombinant Antibody (D9S6V) (CBMAB-CP0017-LY)

-
Mouse Anti-ATP1B1 Recombinant Antibody (E4) (CBMAB-0463-LY)

-
Mouse Anti-CD83 Recombinant Antibody (HB15) (CBMAB-C1765-CQ)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






