METTL3 Antibodies
Background
METTL3 is a methyltransferase widely present in the cell nucleus of eukaryotes, and its core structure consists of a catalytic domain and a zinc finger domain. This enzyme directly regulates the splicing, transport and translation efficiency of messenger RNA by catalyzing the methylation modification (m6A) of the sixth nitrogen atom of adenine in RNA molecules, thereby influencing the gene expression process. As the first confirmed core component of the m6A methyltransferase complex, METTL3 was discovered in 1997. The analysis of its functional mechanism not only reveals important pathways for epigenetic transcriptome regulation but also provides key molecular targets for the study of physiological and pathological processes such as tumorigenesis and stem cell differentiation. In-depth research on this gene continuously drives the cognitive innovation of the RNA epigenetic regulatory network and its mechanism of action in diseases.
Structure of METTL3
METTL3 is a methyltransferase with a molecular weight of approximately 70 kDa. Its precise molecular weight fluctuates within the range of 68-72 kDa due to differences in biological species and transcript isomers.
| Species | Human | Mouse | Zebrafish | Fruit fly |
| Molecular Weight (kDa) | 70.0 | 69.8 | 68.5 | 71.2 |
| Primary Structural Differences | Contains the catalytic MT-A70 domain | Highly homologous to humans | Conservative core structure domain | Simplify the structure of methyl transferase |
This protein is composed of approximately 580 amino acids, and its core function depends on the MT-A70 domain, which forms a unique pocket-like spatial structure to accommodate the S-adenosylmethionine (SAM) cofactor. METTL3 functions by forming a stable heterodimer with METTL14, where METTL3 is responsible for catalyzing reactions. The conserved amino acids in its active center (such as Glu/Asp residues) directly participate in the methyl transfer process, ensuring its specific recognition of RNA and catalysis of N6-methyladenine (m6A) modification.
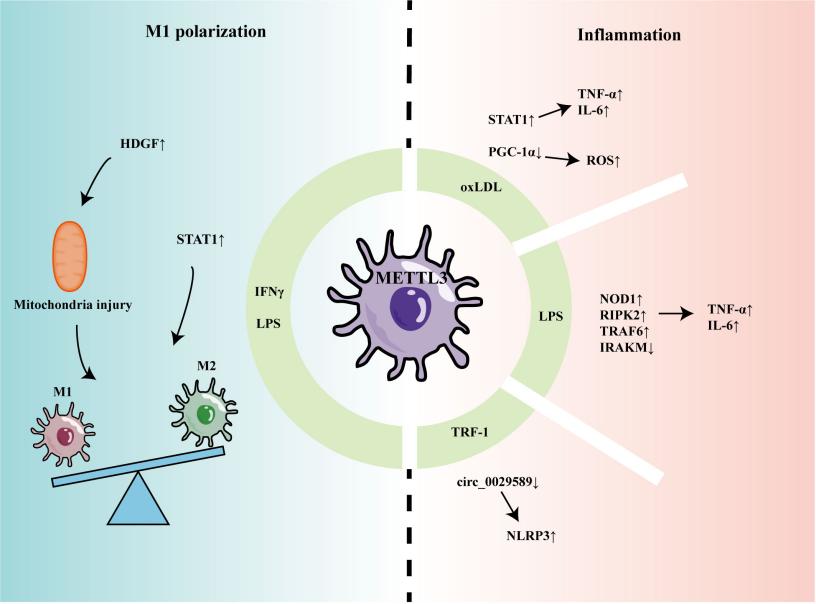 Fig. 1 The role of METTL3 in regulating macrophage inflammation and polarization.1
Fig. 1 The role of METTL3 in regulating macrophage inflammation and polarization.1
Key structural properties of METTL3:
- Classical methyltransferase folding domain (MT-A70 domain)
- Heterodimer interface formed with METTL14
- S-adenosylmethionine (SAM) binding pocket
Functions of METTL3
The core function of METTL3 is to catalyze RNA methylation modification. However, it is also involved in a variety of biological processes such as transcriptional regulation, cell differentiation and tumorigenesis.
| Function | Description |
| m6A methylation catalysis | As the core of the methyltransferase complex, it catalyzes the methylation of the sixth nitrogen atom of adenine on messenger RNA, forming an epigenetic transcriptome marker. |
| Transcriptional regulation | The splicing maturation, nucleoplasmic transport and translation efficiency of pre-mRNA are regulated through m6A modification, which affects the gene expression rate. |
| Cell fate determination | Mediating the maintenance and differentiation process of stem cell pluripotency, its abnormal activity can lead to the disorder of cell fate determination. |
| Tumor-promoting effect | In a wide variety of cancer high expression, by modifying this enhance the stability of cancer gene transcription, promoting tumor proliferation and metastasis. |
| Metabolic regulation | m6A modification of transcripts related to cellular energy metabolism affects metabolic pathways such as glycolysis and oxidative phosphorylation. |
The m6A modification catalyzed by METTL3 exhibits unique "writer" functional characteristics, with its modification distribution highly concentrated near the stop codon of mRNA and in the 3'utr region. This site-specific distribution pattern is closely related to its core role in post-transcriptional regulation.
Applications of METTL3 and METTL3 Antibody in Literature
1. Sun, Yueqin, et al. "METTL3 promotes chemoresistance in small cell lung cancer by inducing mitophagy." Journal of Experimental & Clinical Cancer Research 42.1 (2023): 65. https://doi.org/10.1186/s13046-023-02638-9
Studies have shown that METTL3 promotes chemotherapy resistance in small cell lung cancer by mediating the m6A methylation degradation of DCP2 and activating mitochondrial autophagy in the Pink1-Parkin pathway. Inhibiting METTL3 can reverse this drug resistance process.
2. Wan, Weijun, et al. "METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N6-methyladenosine modification of PD-L1 mRNA in breast cancer." Molecular cancer 21.1 (2022): 60. https://doi.org/10.1186/s12943-021-01447-y
Research has found that METTL3 promotes the degradation of DCP2 through m6A modification, thereby activating mitochondrial autophagy mediated by the Pink1-Parkin pathway, and ultimately leading to chemotherapy resistance in small cell lung cancer. This drug resistance can be reversed by METTL3 inhibitors.
3. Song, Bimei, et al. "Emerging role of METTL3 in inflammatory diseases: mechanisms and therapeutic applications." Frontiers in immunology 14 (2023): 1221609. https://doi.org/10.3389/fimmu.2023.1221609
Research has found that METTL3, as a key m6A methyltransferase, plays a core role in the occurrence and development of various inflammatory diseases by regulating the function of immune cells and the expression of target genes. Natural medicinal components targeting METTL3 offer new strategies for its treatment.
4. Wang, Xiaotong, et al. "METTL3-mediated m6A modification of SIRT1 mRNA inhibits progression of endometriosis by cellular senescence enhancing." Journal of Translational Medicine 21.1 (2023): 407. https://doi.org/10.1186/s12967-023-04209-0
Research has found that METTL3 regulates the stability of SIRT1 mRNA through m6A modification, mediates its degradation through YTHDF2, inhibits the SIRT1/FOXO3a signaling pathway, thereby promoting the senescence of endometrial stem cells and inhibiting their implantation in ectopic areas, which affects the progression of endometriosis.
5. Kang, Ning, et al. "METTL3 regulates thyroid cancer differentiation and chemosensitivity by modulating PAX8." International Journal of Biological Sciences 20.9 (2024): 3426. https://doi.org/10.7150/ijbs.84797
Studies have found that METTL3 exerts a tumor suppressor effect in thyroid cancer, and its expression is negatively regulated by miR-493-5p. METTL3 modifies PAX8 mRNA with m6A and reads it by YTHDC1, inhibiting tumor progression and dedifferentiation, while enhancing the sensitivity of cancer cells to chemotherapy and iodine-131.
Creative Biolabs: METTL3 Antibodies for Research
Creative Biolabs specializes in the production of high-quality METTL3 antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom METTL3 Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our METTL3 antibodies, custom preparations, or technical support, contact us at email.
Reference
- Song, Bimei, et al. "Emerging role of METTL3 in inflammatory diseases: mechanisms and therapeutic applications." Frontiers in immunology 14 (2023): 1221609. https://doi.org/10.3389/fimmu.2023.1221609
Anti-METTL3 antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-AMACR Recombinant Antibody (CB34A) (CBMAB-CA034LY)

-
Mouse Anti-8-oxoguanine Recombinant Antibody (V2-7697) (CBMAB-1869CQ)

-
Mouse Anti-CD2AP Recombinant Antibody (BR083) (CBMAB-BR083LY)

-
Mouse Anti-ARHGAP5 Recombinant Antibody (54/P190-B) (CBMAB-P0070-YC)

-
Mouse Anti-ADRB2 Recombinant Antibody (V2-180026) (CBMAB-A1420-YC)

-
Mouse Anti-2C TCR Recombinant Antibody (V2-1556) (CBMAB-0951-LY)

-
Rabbit Anti-ATF4 Recombinant Antibody (D4B8) (CBMAB-A3872-YC)

-
Mouse Anti-C1QC Recombinant Antibody (CBFYC-0600) (CBMAB-C0654-FY)

-
Mouse Anti-DHFR Recombinant Antibody (D0821) (CBMAB-D0821-YC)

-
Mouse Anti-FLT1 Recombinant Antibody (11) (CBMAB-V0154-LY)

-
Mouse Anti-CORO1A Recombinant Antibody (4G10) (V2LY-1206-LY806)

-
Rabbit Anti-CBL Recombinant Antibody (D4E10) (CBMAB-CP0149-LY)

-
Rabbit Anti-CAMK2A Recombinant Antibody (BA0032) (CBMAB-0137CQ)

-
Mouse Anti-ENO2 Recombinant Antibody (85F11) (CBMAB-0276CQ)

-
Mouse Anti-ENPP1 Recombinant Antibody (CBFYE-0159) (CBMAB-E0375-FY)

-
Mouse Anti-ALOX5 Recombinant Antibody (33) (CBMAB-1890CQ)

-
Mouse Anti-CFL1 (Phospho-Ser3) Recombinant Antibody (CBFYC-1770) (CBMAB-C1832-FY)

-
Mouse Anti-CD247 Recombinant Antibody (6B10.2) (CBMAB-C1583-YY)

-
Mouse Anti-BPGM Recombinant Antibody (CBYY-1806) (CBMAB-2155-YY)

-
Mouse Anti-ACTN4 Recombinant Antibody (V2-6075) (CBMAB-0020CQ)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot








