KMT2A Antibodies
Background
The KMT2A gene encodes a key histone methyltransferase, which is mainly present in the cell nucleus and acts as a transcriptional co-activator to regulate gene expression. This protein activates target genes related to hematopoietic development and cell differentiation by catalyzing the trimethylation modification of lysine 4 at histone H3, maintaining normal hematopoietic stem cell function. In patients with acute leukemia, the generation of the KMT2A fusion gene due to chromosomal translocation can lead to hematopoietic differentiation arrest and malignant proliferation. This discovery originated from the research on the abnormality of chromosome 11q23 in leukemia in the 1990s. Its complex multi-domain conformation provides an important model for the study of epigenetic regulatory mechanisms and profoundly influences the cognitive development in fields such as chromatin modification, transcriptional regulation, and tumorigenesis mechanisms.
Structure of KMT2A
KMT2A is a nucleoprotein with an extremely high molecular weight. The molecular weight of its human isomer is approximately within the range of 430-550 kDa, with specific values varying depending on different splicing variants. This significant molecular weight difference mainly stems from the diversity of protein domain composition and transcript length.
| Species | Human | Mouse | Zebrafish | Fruit fly |
| Molecular Weight (kDa) | 430-550 | ~450 | ~300 | ~200 |
| Primary Structural Differences | Contains multiple functional domains | Highly conservative with humans | Core domain retention | Only basic functional areas are available |
This protein is composed of over 3,500 amino acid residues, and its primary structure contains multiple highly conserved functional modules. The core feature of the KMT2A protein structure is the presence of a catalytically active SET domain, which is responsible for catalyzing the methylation modification of lysine at the 4th position of histone H3. The C-terminal of a protein usually contains a FYRN domain related to transcriptional regulation, while the N-terminal contains multiple XX repeat sequences. These regions jointly mediate protein-protein and protein-DNA interactions. Its tertiary structure is maintained stable through multiple zinc finger molds, forming a functional scaffold capable of recruiting various transcriptional cofactors.
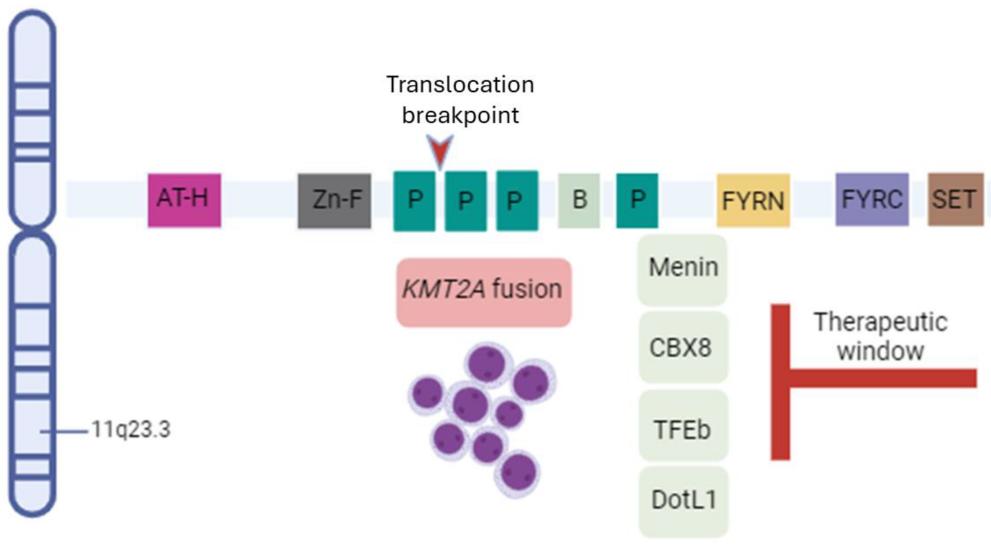 Fig. 1 Schematic representation of KTM2A gene structure and cofactors.1
Fig. 1 Schematic representation of KTM2A gene structure and cofactors.1
Key structural properties of KMT2A:
- Extended conformation composed of multiple domains
- SET domain structure constitute the catalytic activity center
- Multiple zinc finger modalities mediate nucleic acid interactions
Functions of KMT2A
The core function of the KMT2A protein is to regulate gene transcription as a histone methyltransferase. Its main physiological functions and pathological significance are as follows:
| Function | Description |
| Transcriptional activation | Catalyze histone H3K4 methylation, establish an open chromatin structure, and initiate the gene transcription program. |
| Regulation of embryonic development | Guide the maintenance of embryonic stem cell pluripotency and the early development process through epigenetic mechanisms. |
| Maintenance of the hematopoietic system | Regulate the expression of key genes in hematopoietic stem cells to ensure the establishment of normal hematopoietic differentiation lineages. |
| Leukemia occurrence | The KMT2A fusion protein produced by chromosomal translocation leads to hematopoietic differentiation arrest and malignant proliferation. |
| Cell cycle regulation | By influencing the cycle protein dependent kinase inhibitors gene expression involved in cell cycle progression. |
The methylation function of KMT2A depends on its specific binding to nucleosomes. This mode of action is independent of DNA sequence characteristics, reflecting the unique nature of epigenetic regulation and making it a key molecular hub connecting chromatin state and gene expression.
Applications of KMT2A and KMT2A Antibody in Literature
1. Guarnera, Luca, et al. "KMT2A rearrangements in leukemias: molecular aspects and therapeutic perspectives." International journal of molecular sciences 25.16 (2024): 9023. https://doi.org/10.3390/ijms25169023
The article indicates that KMT2A gene rearrangement is more common in childhood leukemia, which can easily lead to chemotherapy resistance and a high risk of recurrence, and the survival rate of patients is generally low. The current treatment strategies include intense chemotherapy, targeted therapy and hematopoietic stem cell transplantation. In particular, transplantation has shown remarkable effects on some children. This article reviews the variant types, clinical features and treatment progress of KMT2A.
2. Ogino, Jayme, and Yali Dou. "Histone methyltransferase KMT2A: Developmental regulation to oncogenic transformation." Journal of Biological Chemistry 300.10 (2024): 107791. https://doi.org/10.1016/j.jbc.2024.107791
The article indicates that KMT2A is an important histone methyltransferase, and its abnormal function is closely related to various diseases, especially leukemia. This article reviews the structure and function of KMT2A and its role in development and tumors, and explores the related targeted therapeutic strategies.
3. Yin, Lei, et al. "Recent Developments and Evolving Therapeutic Strategies in KMT2A‐Rearranged Acute Leukemia." Cancer Medicine 13.20 (2024): e70326. https://doi.org/10.1002/cam4.70326
The article indicates that KMT2A gene rearrangement is a common and high-risk variant in acute leukemia, closely related to strong invasiveness and poor prognosis. This review systematically explores the characteristics, mechanisms and clinical status of KMT2A rearrangement leukemia, and looks forward to new targeted and immunotherapy strategies based on its biological characteristics.
4. Cowell, Ian G., and Caroline A. Austin. "DNA fragility at the KMT2A/MLL locus: insights from old and new technologies." Open Biology 13.1 (2023): 220232. https://doi.org/10.1098/rsob.220232
The article indicates that the KMT2A gene is prone to rearrangement in specific regions, especially after treatment with topoisomerase inhibitors (such as etoposide). Although the drug can induce extensive DNA breaks in this area, the clinically observed hotspots of breaks are relatively concentrated, indicating that in addition to direct cleavage, other factors such as the secondary structure of DNA also jointly determine the selection of break sites.
5. Zhang, Changlin, et al. "KMT2A regulates cervical cancer cell growth through targeting VDAC1." Aging (Albany NY) 12.10 (2020): 9604. https://doi.org/10.18632/aging.103229
The article indicates that KMT2A promotes tumor growth in cervical cancer by regulating VDAC1 expression. Inhibiting KMT2A can impede the proliferation and migration of cancer cells and induce apoptosis. Clinical data also show that KMT2A and VDAC1 are co-highly expressed in cancer tissues. This signal axis provides a new target for the research on the mechanism and treatment of cervical cancer.
Creative Biolabs: KMT2A Antibodies for Research
Creative Biolabs specializes in the production of high-quality KMT2A antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom KMT2A Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our KMT2A antibodies, custom preparations, or technical support, contact us at email.
Reference
- Guarnera, Luca, et al. "KMT2A rearrangements in leukemias: molecular aspects and therapeutic perspectives." International journal of molecular sciences 25.16 (2024): 9023. https://doi.org/10.3390/ijms25169023
Anti-KMT2A antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-CD24 Recombinant Antibody (2Q1282) (CBMAB-C1624-CN)

-
Mouse Anti-ACTB Recombinant Antibody (V2-179553) (CBMAB-A0870-YC)

-
Mouse Anti-AHCYL1 Recombinant Antibody (V2-180270) (CBMAB-A1703-YC)

-
Mouse Anti-ESR1 Recombinant Antibody (Y31) (CBMAB-1208-YC)

-
Mouse Anti-14-3-3 Pan Recombinant Antibody (V2-9272) (CBMAB-1181-LY)

-
Rabbit Anti-DLK1 Recombinant Antibody (9D8) (CBMAB-D1061-YC)

-
Mouse Anti-AKT1 (Phosphorylated S473) Recombinant Antibody (V2-505430) (PTM-CBMAB-0067LY)

-
Mouse Anti-ENO1 Recombinant Antibody (8G8) (CBMAB-E1329-FY)

-
Rabbit Anti-ENO2 Recombinant Antibody (BA0013) (CBMAB-0272CQ)

-
Mouse Anti-CCND2 Recombinant Antibody (DCS-3) (CBMAB-G1318-LY)

-
Mouse Anti-AAV-5 Recombinant Antibody (V2-503416) (CBMAB-V208-1402-FY)

-
Mouse Anti-ENPP1 Recombinant Antibody (CBFYE-0159) (CBMAB-E0375-FY)

-
Mouse Anti-BLNK Recombinant Antibody (CBYY-0623) (CBMAB-0626-YY)

-
Mouse Anti-AOC3 Recombinant Antibody (CBYY-0014) (CBMAB-0014-YY)

-
Mouse Anti-Acetyl-α-Tubulin (Lys40) Recombinant Antibody (V2-623485) (CBMAB-CP2897-LY)

-
Mouse Anti-DLG1 Monolconal Antibody (4F3) (CBMAB-0225-CN)

-
Mouse Anti-CD2AP Recombinant Antibody (BR083) (CBMAB-BR083LY)

-
Mouse Anti-BZLF1 Recombinant Antibody (BZ.1) (CBMAB-AP705LY)

-
Rabbit Anti-BRCA2 Recombinant Antibody (D9S6V) (CBMAB-CP0017-LY)

-
Rabbit Anti-B2M Recombinant Antibody (CBYY-0059) (CBMAB-0059-YY)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






