GATC Antibodies
Background
The GATC gene is widely present in prokaryotes and eukaryotes. The DNA adenine methyltransferase it encodes can catalyze the methylation modification of adenine in specific sequences. This epigenetic marker mainly participates in the regulation of DNA mismatch repair in bacteria, while in eukaryotic cells it affects chromatin structure and gene expression. In the 1980s, researchers first clarified its functional mechanism in Escherichia coli and found that the enzyme distinguishes new DNA strands by recognizing the "GATC" palindrome sequence. As a classic model for DNA methylation research, the GATC gene system not only reveals the fundamental principles of epigenetic regulation but also provides molecular-level explanations for key biological processes such as maintaining genomic stability and regulating the cell cycle.
Structure of GATC
The GATC gene is a key gene encoding DNA adenine methyltransferase in prokaryotes. The molecular weight of its product is approximately 32 kDa, and there is a fluctuation of about ±2 kDa among different strains due to differences in regulatory domains.
| Species | E. coli K12 | Salmonella typhimurium | Klebsiella pneumoniae | Vibrio cholerae | Thermophilic bacteria |
| Gene length (bp) | 834 | 837 | 831 | 846 | 819 |
| Functional features | Replication initiation regulation | Virulence factor modification | Regulation of capsule synthesis | Two-chromosome synchronization | Heat-resistant catalytic domain |
The methyltransferase encoded by this gene specifically recognizes the "GATC" palindrome sequence by forming a tetramer structure. Its active center contains a highly conserved NPPY motif, which can catalyze the transfer of the methyl group of S-adenosylmethionine to the nitrogen atom at position 6 of adenine. This process relies on the synergistic effect of the DNA-binding groove composed of β -sheets and the catalytic core composed of α -helicles in terms of spatial conformation. Among them, the phenylalanine residue at position 119 plays a key role in sequence recognition, while glutamic acid at position 145 directly participates in the methyl transfer reaction.
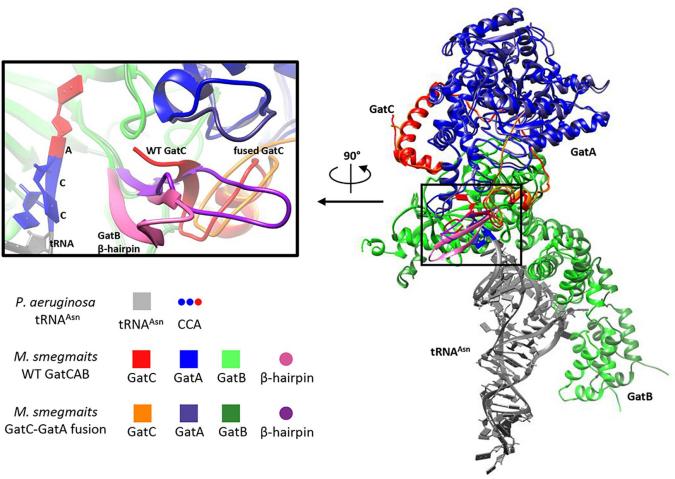 Fig. 1 Modeling suggests GatC-GatA fusion disrupts GatCAB structure.1
Fig. 1 Modeling suggests GatC-GatA fusion disrupts GatCAB structure.1
Key structural properties of GATC:
- Typical palindromic symmetric sequence structure
- Big ditch area presents specific hydrogen bond donor model
- N6 of adenine is the methylation modification site
- Dam methylase achieves specific recognition through conserved domains
Functions of GATC
The core function of the GATC sequence lies in serving as a molecular switch for epigenetic regulation. Its main physiological functions include:
| Function | Description |
| Regulation of DNA replication | Methylation status is used to distinguish parental strand from offspring strand and precisely control the timing of replication initiation. |
| Mismatch repair orientation | Provide chain recognition signals for repair proteins to ensure that incorrect bases are accurately cleared. |
| Regulation of gene expression | The methylation modification of GATC in the promoter region alters the local conformation and affects the binding of transcription factors. |
| Virulence factor management | The random expression of virulence genes is controlled through phase variation mechanisms in pathogenic bacteria. |
| Restricted modification system | As an identification marker of the bacterial self-defense system, it distinguishes itself from foreign DNA. |
The methylation regulation of this sequence presents a dynamic equilibrium feature, and its modification cycle is highly synchronized with the bacterial cell cycle. This temporal characteristic makes it a key hub connecting epigenetic information with the physiological state of cells.
Applications of GATC and GATC Antibody in Literature
1. Du, Chao, et al. "System-wide analysis of the GATC-binding nucleoid-associated protein Gbn and its impact on Streptomyces development." Msystems 7.3 (2022): e00061-22. https://doi.org/10.1128/msystems.00061-22
In this study, a novel chromosomal associated protein, Gbn, was discovered in actinomycetes, which can specifically recognize the GATC sequence motif. Gbn regulates gene expression extensively by binding to 55% of the GATC sites in the genome, influencing the development of Streptomyces sky-blue and antibiotic synthesis, revealing its new functions in bacterial chromosomal organization and gene regulation.
2. Noutahi, Emmanuel, and Nadia El-Mabrouk. "GATC: a genetic algorithm for gene tree construction under the Duplication-Transfer-Loss model of evolution." BMC genomics 19.Suppl 2 (2018): 102. https://doi.org/10.1186/s12864-018-4455-x
In this study, a novel chromosomal tissue protein, Gbn, was discovered in actinomycetes. It can specifically bind to the GATC sequence and widely regulate gene expression, thereby influencing the development of Streptomyces sky-blue and the yield of antibiotics.
3. Cai, Rong-Jun, et al. "Forward genetics reveals a gatc-gata fusion polypeptide causes mistranslation and rifampicin tolerance in Mycobacterium smegmatis." Frontiers in Microbiology 11 (2020): 577756. https://doi.org/10.3389/fmicb.2020.577756
This study, through genetic screening, found that mutations in the gatCAB gene cluster (encoding the key TrNA-modifying enzyme GatCAB) are the main cause of translation errors in mycobacteria. Among them, the GATC-GATA fusion protein induced by the deletion of the GatC stop codon is a common mutation type. It induces mistranslation by disrupting tRNA recognition and enhances the bacterial tolerance to rifampicin.
4. Mardenborough, Yannicka SN, et al. "The unstructured linker arms of MutL enable GATC site incision beyond roadblocks during initiation of DNA mismatch repair." Nucleic acids research 47.22 (2019): 11667-11680. https://doi.org/10.1093/nar/gkz834
This study reveals the molecular mechanism by which the DNA mismatch repair protein MutL in Escherichia coli transmits signals along the DNA strand through its long disordered linker region, thereby activating GATC site cleavage. Experiments show that even in the presence of protein barriers, this mechanism can still initiate incisions at the distal GATC site.
5. Riva, Alessandra, et al. "Characterization of the GATC regulatory network in E. coli." BMC genomics 5.1 (2004): 48. https://doi.org/10.1186/1471-2164-5-48
This study conducted an in-depth analysis of the gene network regulated by GATC sequence methylation in Escherichia coli. This network mainly regulates energy metabolism, respiration, etc. It may be activated when cells encounter cold shock and enter a stable phase, providing survival advantages for bacteria through mechanisms such as apnea.
Creative Biolabs: GATC Antibodies for Research
Creative Biolabs specializes in the production of high-quality GATC antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom GATC Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our GATC antibodies, custom preparations, or technical support, contact us at email.
Reference
- Cai, Rong-Jun, et al. "Forward genetics reveals a gatc-gata fusion polypeptide causes mistranslation and rifampicin tolerance in Mycobacterium smegmatis." Frontiers in Microbiology 11 (2020): 577756. https://doi.org/10.3389/fmicb.2020.577756
Anti-GATC antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-APC Recombinant Antibody (CBYC-A661) (CBMAB-A3036-YC)

-
Mouse Anti-ALPL Antibody (B4-78) (CBMAB-1009CQ)

-
Mouse Anti-BRCA2 Recombinant Antibody (CBYY-1728) (CBMAB-2077-YY)

-
Mouse Anti-CD63 Recombinant Antibody (CBXC-1200) (CBMAB-C1467-CQ)

-
Mouse Anti-ESR1 Recombinant Antibody (Y31) (CBMAB-1208-YC)

-
Mouse Anti-COL12A1 Recombinant Antibody (CBYY-C3117) (CBMAB-C4560-YY)

-
Mouse Anti-CD19 Recombinant Antibody (CBXC-1224) (CBMAB-C1491-CQ)

-
Mouse Anti-ADGRE5 Recombinant Antibody (V2-360335) (CBMAB-C2088-CQ)

-
Rabbit Anti-ATF4 Recombinant Antibody (D4B8) (CBMAB-A3872-YC)

-
Mouse Anti-AOC3 Recombinant Antibody (CBYY-0014) (CBMAB-0014-YY)

-
Mouse Anti-C1QC Recombinant Antibody (CBFYC-0600) (CBMAB-C0654-FY)

-
Rabbit Anti-ABL1 (Phosphorylated Y245) Recombinant Antibody (V2-505716) (PTM-CBMAB-0465LY)

-
Rabbit Anti-Acetyl-Histone H3 (Lys36) Recombinant Antibody (V2-623395) (CBMAB-CP0994-LY)

-
Mouse Anti-AQP2 Recombinant Antibody (G-3) (CBMAB-A3359-YC)

-
Mouse Anti-EMP3 Recombinant Antibody (CBFYE-0100) (CBMAB-E0207-FY)

-
Mouse Anti-DDC Recombinant Antibody (8E8) (CBMAB-0992-YC)

-
Mouse Anti-ARHGDIA Recombinant Antibody (CBCNA-009) (CBMAB-R0415-CN)

-
Mouse Anti-CD1C Recombinant Antibody (L161) (CBMAB-C2173-CQ)

-
Mouse Anti-A2M Recombinant Antibody (V2-178822) (CBMAB-A0036-YC)

-
Mouse Anti-ARG1 Recombinant Antibody (CBYCL-103) (CBMAB-L0004-YC)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






