MSH6 Antibodies
Background
MSH6 gene encoding a key DNA mismatch repair (MMR) protein, mainly exists in eukaryotic nuclei, as part of the than alpha compounds play a role. This protein specifically recognizes and binds to mismatched bases and insertion/deletion errors generated during DNA replication, initiating repair pathways to maintain genomic stability. Patients with Lynch syndrome often carry germline mutations in the MSH6 gene, which significantly increases their susceptibility to cancer. This gene was first identified in 1997. Its functional research not only revealed the molecular mechanism of hereditary colorectal cancer but also promoted the development of tumor molecular typing and immunotherapy. As a sophisticated regulatory mechanism of molecular switches, the MSH6 protein remains a key research focus in the field of DNA repair, providing a crucial theoretical basis for targeted cancer therapy.
Structure of MSH6
MSH6 is a DNA mismatch repair protein with a molecular weight of approximately 160 kDa, and its size varies slightly among different species. This protein is composed of 1,360 amino acids in humans, forming a typical peptide chain structure.
| Species | Human | Mouse | Bovine | Zebrafish | Fruit fly |
| Molecular Weight (kDa) | 160 | 158 | 161 | 159 | 157 |
| Primary Structural Differences | Contains ATP binding domain and DNA binding domain | The homologous region is highly conserved | There is a slight variation at the carboxyl terminal | The functional domains are similar | Simplify the functional domain |
The primary structure of the MSH6 protein contains multiple conserved functional modalities, and its secondary structure is mainly composed of an α -helix and β -folding heterodimer. The N-terminal domain of a protein is responsible for identifying mismatched bases, while the C-terminal domain drives the repair process by hydrolyzing ATP. Conserved phenylalanine and glutamine residues directly participate in DNA binding, while histidine and asparagine coordinate the assembly and function of the repair complex.
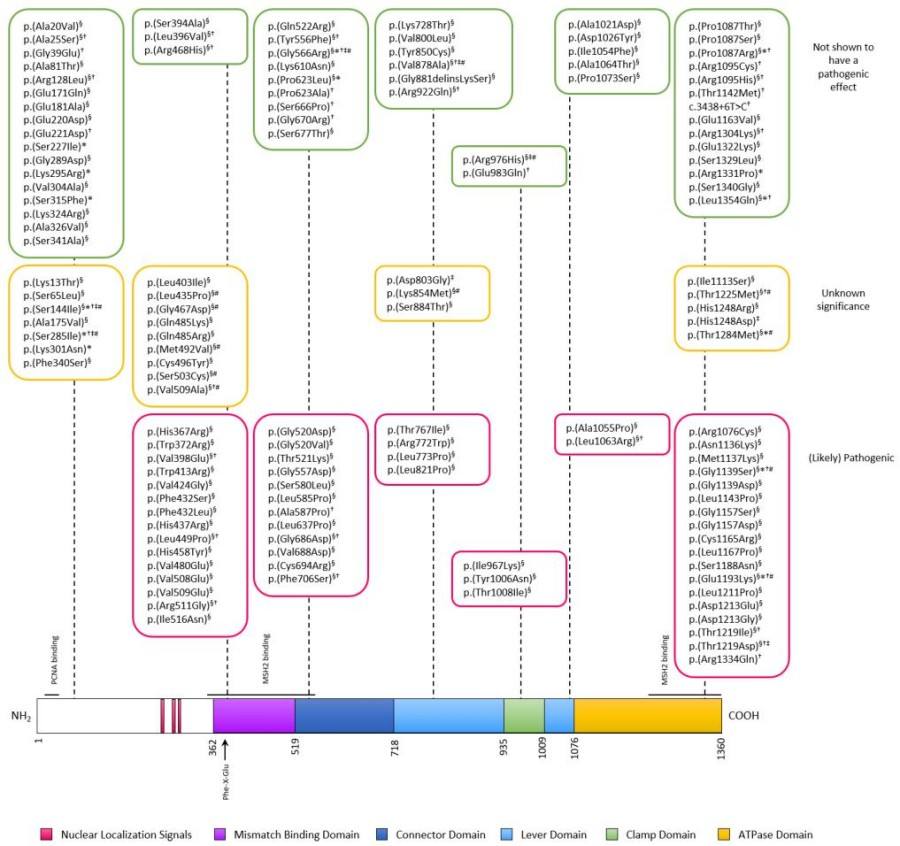 Fig. 1 MSH6 protein domains and analyzed variants.1
Fig. 1 MSH6 protein domains and analyzed variants.1
Key structural properties of MSH6:
- Contains unique DNA binding domains and mismatch recognition segments
- With a conservative ATP hydrolysis combined with functional modules
- Form a stable heterodimer conformation
- The C terminus contains nuclear localization signals and protein interaction regions
- Multiple phosphorylation modification sites are present to regulate repair function
Functions of MSH6
The main function of the MSH6 gene is to participate in DNA mismatch repair to maintain the stability of the genome. In addition, it also involves a variety of biological processes such as cell cycle regulation, apoptosis signal transduction and immune response.
| Function | Description |
| DNA mismatch identification and repair | As a component of the MutSα complex, MSH6 specifically recognizes and binds to DNA replication errors, initiating the repair mechanism. |
| Activation of cell cycle checkpoints | After DNA damage, it participates in blocking the cell cycle process and provides time for repair. |
| Apoptosis induction | In repair DNA damage, failure to start involved in programmed cell death. |
| Regulation of immune diversity | Influence somatic super mutation and antibody type conversion restructuring, which regulates the humoral immune response. |
| Tumor suppressive function | As an important tumor suppressor protein, its functional inactivation is closely related to various hereditary cancers, such as Lynch syndrome. |
The MSH6 protein regulates its binding and dissociation with DNA through its ATPase activity. This characteristic enables it to precisely scan and locate mismatched bases in post-replication complexes. Its functional efficiency directly affects the authenticity of genetic information and the tendency of cells to undergo malignant transformation.
Applications of MSH6 and MSH6 Antibody in Literature
1. Brooksbank, Kirsten, et al. "The DNA mismatch repair protein, MSH6 is a novel regulator of PD-L1 expression." Neoplasia 67 (2025): 101207. https://doi.org/10.1016/j.neo.2025.101207
Studies have found that cancer cells with MSH6 deletion do not show upregulation of PD-L1, which is different from the deletion of MLH1, MSH2, and PMS2. Mechanistically, MSH6 directly regulates PD-L1 transcription through SETD2. MSH6 deletion also leads to a high mutational burden but low microsatellite instability, suggesting that MMR defects need to be evaluated by gene stratification.
2. Wang, Xinyue, et al. "Rescue RM/CS-AKI by blocking strategy with one-dose anti-myoglobin RabMAb." Nature Communications 16.1 (2025): 1044. https://doi.org/10.18632/aging.205329
Studies have found that the expression of long non-coding RNA PTCHD4-AS is down-regulated in gastric cancer. Its up-regulation can promote ATM phosphorylation and activate the p53-p21 pathway by directly binding to the MSH2-MSH6 dimer, thereby inhibiting cell proliferation, enhancing cisplatin sensitivity, and inducing G2/M phase arrest and apoptosis.
3. Danquah, Bright D., et al. "Mass Spectrometric analysis of antibody—Epitope peptide complex dissociation: Theoretical concept and practical procedure of binding strength characterization." Molecules 25.20 (2020): 4776. https://doi.org/10.3390/ijms21082831
Studies have found that MSH6 is lowly expressed in invasive and specific subtypes of non-functional pituitary adenomas, and is positively correlated with PD-L1 expression. Knockout of MSH6 significantly reduces the level of PD-L1 and promotes tumor proliferation, indicating that the loss of its expression may exacerbate the invasiveness and malignant progression of pituitary adenomas.
4. Wang, Lin, et al. "Mismatch Repair Protein Msh6Tt Is Necessary for Nuclear Division and Gametogenesis in Tetrahymena thermophila." International Journal of Molecular Sciences 24.24 (2023): 17619. https://doi.org/10.3390/ijms242417619
The article indicates that in Tetrahymena, Msh6Tt is located in the large and small nuclei and participates in amitosis, meiosis and gametogenesis. Knockout of MSH6Tt leads to abnormal nuclear division, MMS tolerance, and affects the stability of MSH2Tt and the upregulation of other homologous gene expressions.
5. Frederiksen, Jane H., et al. "Classification of MSH6 variants of uncertain significance using functional assays." International Journal of Molecular Sciences 22.16 (2021): 8627. https://doi.org/10.3390/ijms22168627
The article indicates that Lynch syndrome (LS) is a common hereditary cancer susceptibility syndrome caused by pathogenic variations in mismatch repair genes (such as MSH6). This article focuses on reviewing the functional detection methods of MSH6 gene variations and their applications in clinical classification, and explores the development prospects of high-throughput functional analysis.
Creative Biolabs: MSH6 Antibodies for Research
Creative Biolabs specializes in the production of high-quality MSH6 antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom MSH6 Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our MSH6 antibodies, custom preparations, or technical support, contact us at email.
Reference
- Frederiksen, Jane H., et al. "Classification of MSH6 variants of uncertain significance using functional assays." International Journal of Molecular Sciences 22.16 (2021): 8627. https://doi.org/10.3390/ijms22168627
Anti-MSH6 antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-DMPK Recombinant Antibody (CBYCD-324) (CBMAB-D1200-YC)

-
Mouse Anti-CD24 Recombinant Antibody (2Q1282) (CBMAB-C1624-CN)

-
Mouse Anti-CFL1 Recombinant Antibody (CBFYC-1771) (CBMAB-C1833-FY)

-
Rabbit Anti-DLK1 Recombinant Antibody (9D8) (CBMAB-D1061-YC)

-
Mouse Anti-A2M Recombinant Antibody (V2-178822) (CBMAB-A0036-YC)

-
Mouse Anti-CRTAM Recombinant Antibody (CBFYC-2235) (CBMAB-C2305-FY)

-
Mouse Anti-CAPZB Recombinant Antibody (CBYY-C0944) (CBMAB-C2381-YY)

-
Mouse Anti-CTNND1 Recombinant Antibody (CBFYC-2414) (CBMAB-C2487-FY)

-
Mouse Anti-BrdU Recombinant Antibody (IIB5) (CBMAB-1038CQ)

-
Rabbit Anti-Acetyl-Histone H3 (Lys36) Recombinant Antibody (V2-623395) (CBMAB-CP0994-LY)

-
Mouse Anti-CD24 Recombinant Antibody (SN3) (CBMAB-C1037-CQ)

-
Mouse Anti-CD1C Recombinant Antibody (L161) (CBMAB-C2173-CQ)

-
Rabbit Anti-AP2M1 (Phosphorylated T156) Recombinant Antibody (D4F3) (PTM-CBMAB-0610LY)

-
Mouse Anti-CRYAB Recombinant Antibody (A4345) (CBMAB-A4345-YC)

-
Mouse Anti-ATG5 Recombinant Antibody (9H197) (CBMAB-A3945-YC)

-
Mouse Anti-ABCA3 Recombinant Antibody (V2-178911) (CBMAB-A0145-YC)

-
Mouse Anti-CECR2 Recombinant Antibody (CBWJC-2465) (CBMAB-C3533WJ)

-
Mouse Anti-CCDC25 Recombinant Antibody (CBLC132-LY) (CBMAB-C9786-LY)

-
Mouse Anti-DES Monoclonal Antibody (440) (CBMAB-AP1857LY)

-
Mouse Anti-ACTG1 Recombinant Antibody (V2-179597) (CBMAB-A0916-YC)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






