ssDNA Antibodies
Background
Single-stranded DNA (ssDNA) is genetic material existing in the form of a single nucleotide chain, mainly found in the gene expression processes of certain viruses, bacteriophages and eukaryotes. It participates in gene replication, recombination and repair through base complementary pairing, maintaining the stability and diversity of genetic information. The field of gene therapy relies on artificially designed ssDNA for precise gene editing, as its programmability can target specific genomic sequences. Since the clarification of the DNA double helix structure in the 1950s, ssDNA, as a key carrier for genetic regulation, has been continuously and deeply studied. Its efficient and flexible characteristics have greatly promoted the development of molecular diagnosis, nucleic acid drugs and synthetic biology, providing an important tool for biomedical research.
Structure of ssDNA
ssDNA is a nucleic acid polymer with a relatively small molecular weight, typically ranging from several thousand to several hundred thousand daltons. The specific value varies depending on the chain length and base composition. There are certain differences in the molecular weight of ssDNA from different sources:
| Species | Viral ssDNA (M13mp18) | PCR primer (27-mer) | ssDNA aptamer | Synthetic oligonucleotides |
| Molecular Weight (kDa) | ~32.5 | ~8.3 | ~15.6 | According to the sequence |
| Primary Structural Differences | Ring, genome | Linear, specific primers | With a specific three-dimensional conformation | Can be designed and modified |
ssDNA is composed of a nucleotide chain linked by a phosphodiester bond, and its primary structure is based on the order of base arrangement, which determines the hybridization specificity. Molecules can form secondary structures such as hairpins and stem rings, and these folding patterns are crucial for their recognition and binding to target molecules. ssDNA achieves specific binding to target sequences through base complementary pairing, a property that is widely applied in gene editing, molecular probes, and amplification techniques. Its biological function does not require the participation of cofactors, and its stability mainly depends on sequence length, GC content and environmental conditions.
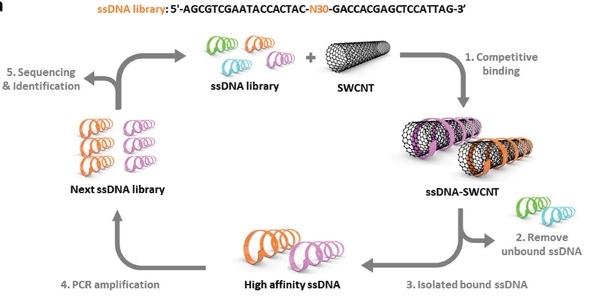 Fig. 1Schematic representation of the selection process for high‐affinity ssDNA sequences on SWCNT surfaces.1
Fig. 1Schematic representation of the selection process for high‐affinity ssDNA sequences on SWCNT surfaces.1
Key structural properties of ssDNA:
- Linear nucleotide single-stranded structure
- Stability is maintained through base stacking force and hydrogen bonds
- Targeted binding and recognition are achieved through base complementary pairing
Functions of ssDNA
The core function of ssDNA is to serve as a carrier of genetic information and a medium for gene regulation, and it also plays a key role in various molecular biological processes.
| Function | Description |
| Genetic information transmission | As the genetic material of certain viruses and bacteriophages, it carries coding genes and completes replication and expression. |
| The function of the template | In DNA replication, PCR amplification and sequencing reaction as a template, and guide the combination of the complementary strand. |
| Molecular recognition and binding | Combining complementary base pairing and target sequence specificity, are widely used in the probe, primers and gene editing tools. |
| Regulation of gene expression | Regulate the expression level of target genes in the form of antisense oligonucleotides or RNA interference precursors. |
| Structural assembly function | As a self-assembly unit in nucleic acid nanotechnology, it is used to construct fine 2D or 3D structures. |
Compared with double-stranded DNA, ssDNA has higher flexibility and can participate in reactions without unwinding, demonstrating faster hybridization kinetics and good programmability, making it an important tool in molecular diagnostics, synthetic biology and therapeutics.
Applications of ssDNA and ssDNA Antibody in Literature
1. Lee, Dakyeon, et al. "Systematic Selection of High‐Affinity ssDNA Sequences to Carbon Nanotubes." Advanced Science 11.32 (2024): 2308915. https://doi.org/10.1002/advs.202308915
In this study, ssDNA sequences with high affinity for single-walled carbon nanotubes were obtained through high-throughput screening, revealing their key base compositions and hydrogen bond interaction mechanisms. A predictive model was constructed by combining molecular simulation and machine learning, providing a new strategy for the design of stable and sensitive biosensors.
2. Liang, Chih-Chao, et al. "Mechanism of single-stranded DNA annealing by RAD52–RPA complex." Nature 629.8012 (2024): 697-703. https://doi.org/10.1038/s41586-024-07347-7
This study reveals the molecular mechanism by which RAD52 binds to RPA by forming an open-loop structure, driving the annealing of complementary ssDNA. Structural analysis indicates that ssDNA is located in the positively charged channel of the RAD52 ring, with its N-terminal domain promoting annealing and the C-terminal regulating the open-ring conformation and RPA interaction. This discovery provides new insights into targeting RAD52 for the treatment of homologous recombination deficiency cancers.
3. Park, Jee-Woong, et al. "Acousto-microfluidics for screening of ssDNA aptamer." Scientific reports 6.1 (2016): 27121. https://doi.org/10.1038/srep27121
In this study, acoustic fluid screening technology was employed in combination with high-throughput sequencing to successfully obtain ssDNA aptamers that bind to prostate-specific antigen (PSA) with high affinity. The dissociation constant of the best candidate molecule reaches 0.7 nM. This method does not require the traditional washing step, significantly improving the screening efficiency and specificity.
4. Okorokov, Andrei L., et al. "Structure of the hDmc1-ssDNA filament reveals the principles of its architecture." PloS one 5.1 (2010): e8586. https://doi.org/10.1371/journal.pone.0008586
In this study, the structure of the pre-synaptic complex formed by human Dmc1 protein and ssDNA was resolved by single-particle electron microscopy. Unlike the RecA/Rad51 family, hDmc1 forms an extended active filament with 9 subunits per circle on ssDNA, each subunit covering two nucleotides, and its N-terminal domain coordinates the role of adjacent organisms through domain exchange mechanisms.
5. Ribeiro, Jonathan, et al. "RPA homologs and ssDNA processing during meiotic recombination." Chromosoma 125.2 (2016): 265-276. https://doi.org/10.1007/s00412-015-0552-7
The article indicates that in meiosis homologous recombination, the formation and maturation of ssDNA are key links. Unlike mitosis, MEIOB and multiple RPA subunits work in synergy to specifically regulate ssDNA processing, ensuring that homologous chromosomes preferentially serve as repair templates, thereby maintaining genomic stability and genetic diversity.
Creative Biolabs: ssDNA Antibodies for Research
Creative Biolabs specializes in the production of high-quality ssDNA antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for PCR, gene editing, sequencing, and nucleic acid hybridization, and other diagnostic methodologies.
- Custom ssDNA Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our ssDNA antibodies, custom preparations, or technical support, contact us at email.
Reference
- Lee, Dakyeon, et al. "Systematic Selection of High‐Affinity ssDNA Sequences to Carbon Nanotubes." Advanced Science 11.32 (2024): 2308915. https://doi.org/10.1002/advs.202308915
Anti-ssDNA antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-ASB9 Recombinant Antibody (1D8) (CBMAB-A0529-LY)

-
Mouse Anti-ACTG1 Recombinant Antibody (V2-179597) (CBMAB-A0916-YC)

-
Mouse Anti-Acetyl SMC3 (K105/K106) Recombinant Antibody (V2-634053) (CBMAB-AP052LY)

-
Mouse Anti-ATP1A2 Recombinant Antibody (M7-PB-E9) (CBMAB-A4013-YC)

-
Mouse Anti-ACE2 Recombinant Antibody (V2-179293) (CBMAB-A0566-YC)

-
Rabbit Anti-Acetyl-Histone H3 (Lys36) Recombinant Antibody (V2-623395) (CBMAB-CP0994-LY)

-
Mouse Anti-BIRC3 Recombinant Antibody (315304) (CBMAB-1214-CN)

-
Mouse Anti-BRD3 Recombinant Antibody (CBYY-0801) (CBMAB-0804-YY)

-
Rabbit Anti-ATF4 Recombinant Antibody (D4B8) (CBMAB-A3872-YC)

-
Human Anti-SARS-CoV-2 S1 Monoclonal Antibody (CBFYR-0120) (CBMAB-R0120-FY)

-
Mouse Anti-FN1 Monoclonal Antibody (D6) (CBMAB-1240CQ)

-
Mouse Anti-AP4E1 Recombinant Antibody (32) (CBMAB-A2996-YC)

-
Mouse Anti-ALB Recombinant Antibody (V2-180650) (CBMAB-A2186-YC)

-
Mouse Anti-APOE Recombinant Antibody (A1) (CBMAB-0078CQ)

-
Mouse Anti-CASP8 Recombinant Antibody (CBYY-C0987) (CBMAB-C2424-YY)

-
Mouse Anti-BLNK Recombinant Antibody (CBYY-0623) (CBMAB-0626-YY)

-
Mouse Anti-ATP1B3 Recombinant Antibody (1E9) (CBMAB-A4021-YC)

-
Rabbit Anti-AKT3 Recombinant Antibody (V2-12567) (CBMAB-1057-CN)

-
Mouse Anti-CCDC6 Recombinant Antibody (CBXC-0106) (CBMAB-C5397-CQ)

-
Mouse Anti-ATG5 Recombinant Antibody (9H197) (CBMAB-A3945-YC)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






