CEBPA Antibodies
Background
The CEBPA gene encodes a protein belonging to the leucine zipper transcription factor family and is mainly expressed in bone marrow hematopoietic tissue. This protein promotes the differentiation and maturation of myeloid cells and maintains hematopoietic homeostasis by regulating the expression of downstream target genes. In hematological malignancies such as acute myeloid leukemia, mutations in the CEBPA gene often lead to its loss of function, which in turn affects normal myeloid differentiation and promotes malignant proliferation. This gene was first identified in the early 1990s. The study of its structure and function has provided a key foundation for understanding the mechanism of hematopoietic differentiation and the pathogenesis of leukemia. CEBPA, as a core regulatory factor of myeloid differentiation, has become an important object in the research of molecular diagnosis, prognosis evaluation and targeted therapy of leukemia.
Structure of CEBPA
The molecular weight of the transcription factor protein encoded by the CEBPA gene is approximately 35 kDa. There are subtle differences in the molecular weight of this protein among different species, mainly due to the species specific variations in the sequences of its N-terminal regulatory domain and C-terminal basic leucine zipline (bZIP) domain.
| Species | Human | Mouse | Rat |
| Molecular Weight (kDa) | About 35 | About 34 | About 35 |
| Primary Structural Differences | There are various isomers produced at the beginning of translation | The core functional area is highly conservative | High homology with human protein sequence |
The core structure of the CEBPA protein is a highly conserved C-terminal basic leucine zipper (bZIP) domain, which is responsible for mediating protein dimerization and binding to specific DNA sequences, thereby regulating the transcription of downstream target genes. Its N-terminal contains a transcriptional activation domain, but the sequence and length of this region vary greatly among different species or isomers, which directly affects the fineness of its transcriptional activity and functional regulation. This protein usually functions in the form of a homodimer, and its structural characteristics are the molecular basis for it to perform the function of a "master regulatory factor" in myeloid differentiation.
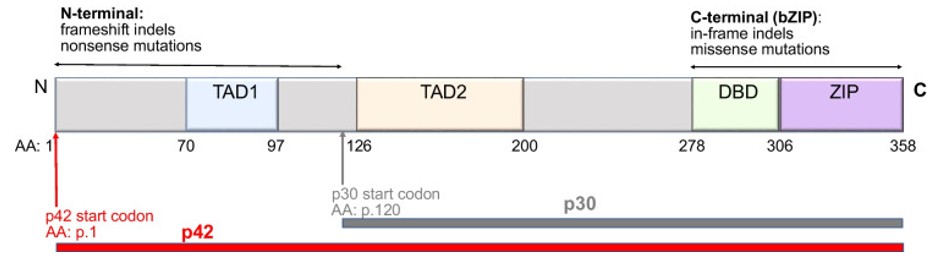 Fig. 1 Schematic of the CEBPA protein structure and distribution of mutations.1
Fig. 1 Schematic of the CEBPA protein structure and distribution of mutations.1
Key structural properties of CEBPA:
- Conserved basic leucine zipper (bZIP) dimerization binds to the DNA domain
- Flexible N-terminal transcriptional activation domain
- Specific homology/heterodimer interfaces formed through leucine zips
Functions of CEBPA
The main function of the CEBPA gene-encoded protein is to act as a transcription factor to regulate the differentiation and proliferation of myeloid cells. However, it is also widely involved in a variety of cellular physiological processes, including metabolic reprogramming, cell cycle regulation and tumor suppression.
| Function | Description |
| Regulation of myeloid differentiation | Activate the expression of a series of granulocyte-specific genes to drive the differentiation of hematopoietic stem cells into mature granulocytes. |
| Cell cycle arrest | By up-regulating genes such as p21 and inhibiting cyclin-dependent kinases, it promotes cell exit from the cell cycle. |
| Metabolic reprogramming | Regulating genes related to mitochondrial function and glucose metabolism affects the energy metabolism state of cells. |
| Tumor suppression | In acute myeloid leukemia, its loss-of-function mutations lead to differentiation arrest and malignant proliferation, playing an important role in tumor suppression. |
| Maintenance of hematopoietic homeostasis | With other transcription factors (such as PU. 1) together, fine balance of self-renewal and differentiation of hematopoietic progenitor cells. |
CEBPA usually binds to DNA in the form of a homodimer, and its transcriptional activation kinetics shows a synergistic relationship rather than a simple linear one. This enables it to act as a "molecular switch" for determining cell fate and play an irreplaceable regulatory role at key nodes of hematopoietic differentiation.
Applications of CEBPA and CEBPA Antibody in Literature
1. Georgi, Julia-Annabell, et al. "Prognostic impact of CEBPA mutational subgroups in adult AML." Leukemia 38.2 (2024): 281-290. https://doi.org/10.1038/s41375-024-02140-x
This study analyzed the data of 1,010 adult AML patients with CEBPA-mutant mutations and found that patients with only bZIP in-box insertion/deletion mutations (bZIPInDel) had a higher complete response rate and longer survival period, and the prognosis of this subtype was not affected by other co-mutations. The results suggest that the current classification standards of WHO, ELN2022 and ICC need to be further refined.
2. Cai, Yunshi, et al. "LncRNA CEBPA-DT promotes liver cancer metastasis through DDR2/β-catenin activation via interacting with hnRNPC." Journal of Experimental & Clinical Cancer Research 41.1 (2022): 335. https://doi.org/10.1186/s13046-022-02544-6
Research has found that long non-coding RNA CEBPA-DT in hepatocellular carcinoma upregulates DDR2 by binding to hnRNPC and activates the β-catenin/Snail1 pathway to promote tumor metastasis. This axis can serve as a potential therapeutic target.
3. Yuan, Ji, Rong He, and Hassan B. Alkhateeb. "Sporadic and familial acute myeloid leukemia with CEBPA mutations." Current Hematologic Malignancy Reports 18.5 (2023): 121-129. https://doi.org/10.1007/s11899-023-00699-3
This article reviews the classification update of CEBPA-mutated AML, elaborates on its significance for laboratory assessment and clinical management, and discusses risk stratification and transplantation selection. New developments have changed the definition, identification and treatment strategies of this disease.
4. Heyes, Elizabeth, et al. "TET2 lesions enhance the aggressiveness of CEBPA-mutant acute myeloid leukemia by rebalancing GATA2 expression." Nature Communications 14.1 (2023): 6185. https://doi.org/10.1038/s41467-023-41927-x
Research has found that in AML with CEBPA double mutations, the high expression of CEBPA caused by N-terminal mutations and the deletion of TET2 synergically regulate the level of GATA2: CEBPA recruits TET2 to maintain the activity of the Gata2 enhancer, while the deletion of TET2 inhibits its expression through methylation. Demethylation therapy can restore Gata2 levels and delay disease progression.
5. Cadefau-Fabregat, Maria, et al. "Mutant CEBPA promotes tolerance to inflammatory stress through deficient AP-1 activation." Nature Communications 16.1 (2025): 3492. https://doi.org/10.1038/s41467-025-58712-7
Research has found that the p30 protein produced by CEBPA mutations in AML regulates the AP-1 factor differently, reduces the expression of inflammatory genes and alters the stress response, thereby enhancing the cells' tolerance to inflammation and ER stress. This may become a potential therapeutic target.
Creative Biolabs: CEBPA Antibodies for Research
Creative Biolabs specializes in the production of high-quality CEBPA antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom CEBPA Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our CEBPA antibodies, custom preparations, or technical support, contact us at email.
Reference
- Yuan, Ji, Rong He, and Hassan B. Alkhateeb. "Sporadic and familial acute myeloid leukemia with CEBPA mutations." Current Hematologic Malignancy Reports 18.5 (2023): 121-129. https://doi.org/10.1007/s11899-023-00699-3
Anti-CEBPA antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-CD247 Recombinant Antibody (6B10.2) (CBMAB-C1583-YY)

-
Rat Anti-(1-5)-α-L-Arabinan Recombinant Antibody (V2-501861) (CBMAB-XB0003-YC)

-
Mouse Anti-CDKL5 Recombinant Antibody (CBFYC-1629) (CBMAB-C1689-FY)

-
Mouse Anti-CRTAM Recombinant Antibody (CBFYC-2235) (CBMAB-C2305-FY)

-
Rabbit Anti-ABL1 (Phosphorylated Y185) Recombinant Antibody (V2-443434) (PTM-CBMAB-0001YC)

-
Rat Anti-ADAM10 Recombinant Antibody (V2-179741) (CBMAB-A1103-YC)

-
Mouse Anti-APCS Recombinant Antibody (CBYC-A663) (CBMAB-A3054-YC)

-
Mouse Anti-14-3-3 Pan Recombinant Antibody (V2-9272) (CBMAB-1181-LY)

-
Mouse Anti-DHFR Recombinant Antibody (D0821) (CBMAB-D0821-YC)

-
Mouse Anti-AQP2 Recombinant Antibody (G-3) (CBMAB-A3359-YC)

-
Mouse Anti-BIRC3 Recombinant Antibody (315304) (CBMAB-1214-CN)

-
Mouse Anti-AP4E1 Recombinant Antibody (32) (CBMAB-A2996-YC)

-
Rabbit Anti-BAD (Phospho-Ser136) Recombinant Antibody (CAP219) (CBMAB-AP536LY)

-
Mouse Anti-CD24 Recombinant Antibody (ALB9) (CBMAB-0176CQ)

-
Mouse Anti-CA9 Recombinant Antibody (CBXC-2079) (CBMAB-C0131-CQ)

-
Mouse Anti-CAT Recombinant Antibody (724810) (CBMAB-C8431-LY)

-
Mouse Anti-AK4 Recombinant Antibody (V2-180419) (CBMAB-A1891-YC)

-
Rat Anti-EMCN Recombinant Antibody (28) (CBMAB-E0280-FY)

-
Mouse Anti-CD24 Recombinant Antibody (HIS50) (CBMAB-C10123-LY)

-
Mouse Anti-dsRNA Recombinant Antibody (2) (CBMAB-D1807-YC)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot








