NAT1 Antibodies
Background
The NAT1 gene encodes N-acetyltransferase 1 and is mainly present in the liver and intestinal tissues of vertebrates. This gene is responsible for catalyzing the acetylation reaction of aromatic amine compounds, participating in the metabolic detoxification of exogenous substances and the regulation of endogenous bioactive molecules. Individual differences in drug metabolism are often associated with NAT1 gene polymorphisms, and changes in its activity can affect drug efficacy and the risk of toxic and side effects. This gene was first cloned and located in the 1990s and is the first key enzyme gene in the human acetylation metabolic pathway to complete functional analysis. Its highly conserved protein domains and diverse single nucleotide polymorphisms have become important research models in pharmacogenomics, providing a molecular basis for individualized medication and disease susceptibility studies.
Structure of NAT1
The molecular weight of the protein encoded by the NAT1 gene is approximately 34 kDa. There are minor differences in this molecular weight among different species, which is mainly due to variations in amino acid sequences.
| Species | Human | Mouse | Rat |
| Molecular Weight (kDa) | 34.0 | 33.8 | 33.5 |
| Primary Structural Differences | Catalyzing the acetylation of aromatic amines and hydrazine compounds plays a core role in drug metabolism and detoxification | Highly homologous in function, it is a commonly used model for studying drug metabolism | Active sites are highly conserved and are often used in toxicological studies |
N-acetyltransferase 1 (NAT1) is composed of 290 amino acids and forms a stable homodimer structure. The core of its three-dimensional structure is A highly conserved catalytic domain that can bind to acetyl-coenzyme A as a cofactor. This enzyme protein exhibits a characteristic "cloverleaf" folding, and its active center contains a key trivalent residue (Cys-His-Asp), in which the cysteine residue directly participates in the acetyl transfer reaction. Its substrate-binding pocket has a flexible hydrophobic environment, which can specifically recognize and catalyze the N-acetylation of various aromatic amine substances. This structural feature determines its wide substrate spectrum and metabolic differences among individuals.
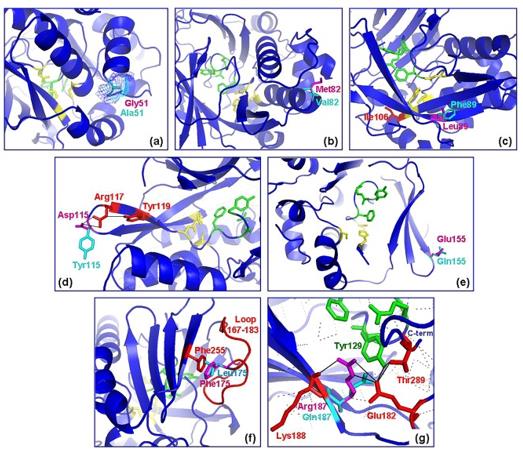 Fig. 1 Position of polymorphic residues on (MACMU)NAT1 protein structure.1
Fig. 1 Position of polymorphic residues on (MACMU)NAT1 protein structure.1
Key structural properties of NAT1:
- Form a stable homodimer structure
- Core is conservative catalytic domain structure, contain hydrophobic substrates combination pocket
- Active center rely on key triplet residues (Cys - His - Asp) shift in acetyl groups
Functions of NAT1
The main function of the NAT1 gene is to catalyze the N-acetylation reaction of aromatic amines and hydrazine compounds, and participate in the drug metabolism and detoxification process of the body. At the same time, it also involves multiple physiological and pathological processes, including the regulation of cancer susceptibility and hormone metabolism.
| Function | Description |
| Drug metabolism | Catalyzing the acetylation inactivation or conversion of a variety of drugs, such as isoniazid and aminosalicylic acid, is a key enzyme that determines the half-life of drugs and differences in individual efficacy. |
| Detoxification effect | The deactivation of exogenous aromatic amine carcinogens by acetylation reduces their genotoxicity and carcinogenic risk. |
| Hormone regulation | Participate in acetylated metabolism of folic acid and its derivatives, and influence of one carbon unit in the cell metabolic balance. |
| Disease association | The gene polymorphism of NAT1 is associated with the susceptibility to diseases such as bladder cancer and colorectal cancer, and changes in its activity can affect an individual's risk of disease when exposed to specific environments. |
| The basis of individualized medication | The genotype (fast/slow acetylated type) is one of the important pharmacological basis for clinical implementation of individualized medication, used to predict drug response and toxicity. |
Unlike the Mie kinetic characteristics of most metabolic enzymes, NAT1 exhibits substrate inhibition on certain substrates, meaning that the reaction rate actually decreases at high concentrations of the substrate. This unique kinetic property suggests that it may have physiological regulatory significance in preventing excessive accumulation of toxic intermediate products within cells, and that its metabolic function is not only determined by the quantity of enzymes but also precisely regulated by the substrate concentration and microenvironment.
Applications of NAT1 and NAT1 Antibody in Literature
- Gu, Wang, et al. "NAT1 inhibits liver metastasis of colorectal cancer by regulating EMT and glycolysis." Aging (Albany NY) 16.12 (2024): 10546. https://doi.org/10.18632/aging.205957
This study confirmed that overexpression of NAT1 inhibits the PI3K/AKT/mTOR pathway, down-regulates the EMT process, glycolytic ability and VEGF expression of colorectal cancer cells, thereby suppressing tumor invasion and metastasis as well as liver microangiogenesis, and ultimately reducing the occurrence of liver metastasis from colorectal cancer.
- Carlisle, Samantha M., et al. "Human arylamine N-acetyltransferase 1 (NAT1) knockout in MDA-MB-231 breast cancer cell lines leads to transcription of NAT2." Frontiers in pharmacology 12 (2022): 803254. https://doi.org/10.3389/fphar.2021.803254
This study found through RNA-seq analysis that changes in the expression level of NAT1 significantly affected the global gene expression of breast cancer MDA-MB-231 cells, involving pathways such as cell adhesion. Moreover, NAT1 knockout could induce the transcription of isoenzyme NAT2, suggesting that its function goes beyond classical drug metabolism.
- Habil, Mariam R., Mark A. Doll, and David W. Hein. "Acetyl coenzyme A kinetic studies on N-acetylation of environmental carcinogens by human N-acetyltransferase 1 and its NAT1* 14B variant." Frontiers in Pharmacology 13 (2022): 931323. https://doi.org/10.3389/fphar.2022.931323
This study reveals that the acetylation metabolic capacity of the NAT1*14B allele for various aromatic amine carcinogens is significantly lower than that of the reference allele NAT1*4, and it has a lower affinity for the cofactor AcCoA when metabolizing BNA and benzidine. This provides a molecular basis for explaining the differences in carcinogen metabolism among individuals.
- Boukouvala, Sotiria, et al. "Population variability of rhesus macaque (Macaca mulatta) NAT1 gene for arylamine N-acetyltransferase 1: Functional effects and comparison with human." Scientific Reports 9.1 (2019): 10937. https://doi.org/10.1038/s41598-019-47485-x
Through the study of the NAT1 gene in rhesus monkeys, it was found that the functional polymorphism within the population is much higher than that in humans. Multiple non-synonymous variations can significantly weaken the enzyme activity, suggesting that the two species may face different environmental pressures. This provides a comparative perspective for studying the functional evolution of NAT1 and its association with diseases.
- Matejcic, Marco, et al. "NAT1 and NAT2 genetic polymorphisms and environmental exposure as risk factors for oesophageal squamous cell carcinoma: a case-control study." BMC cancer 15.1 (2015): 150. https://doi.org/10.1186/s12885-015-1105-4
This study indicates that the slow/moderate metabolisms of NAT1 and NAT2 significantly increase the risk of esophageal cancer in the South African population who smoke or drink alcohol, and interact with the intake of red meat/white meat. Acetylation polymorphisms and environmental factors synergistically affect the risk of canceration.
Creative Biolabs: NAT1 Antibodies for Research
Creative Biolabs specializes in the production of high-quality NAT1 antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom NAT1 Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our NAT1 antibodies, custom preparations, or technical support, contact us at email.
Reference
- Boukouvala, Sotiria, et al. "Population variability of rhesus macaque (Macaca mulatta) NAT1 gene for arylamine N-acetyltransferase 1: Functional effects and comparison with human." Scientific Reports 9.1 (2019): 10937. https://doi.org/10.1038/s41598-019-47485-x
Anti-NAT1 antibodies
 Loading...
Loading...
Hot products 
-
Rabbit Anti-CCL5 Recombinant Antibody (R0437) (CBMAB-R0437-CN)

-
Rabbit Anti-AP2M1 (Phosphorylated T156) Recombinant Antibody (D4F3) (PTM-CBMAB-0610LY)

-
Mouse Anti-ACE2 Recombinant Antibody (V2-179293) (CBMAB-A0566-YC)

-
Mouse Anti-Acetyl SMC3 (K105/K106) Recombinant Antibody (V2-634053) (CBMAB-AP052LY)

-
Mouse Anti-APP Recombinant Antibody (DE2B4) (CBMAB-1122-CN)

-
Mouse Anti-AZGP1 Recombinant Antibody (CBWJZ-007) (CBMAB-Z0012-WJ)

-
Mouse Anti-CD83 Recombinant Antibody (HB15) (CBMAB-C1765-CQ)

-
Mouse Anti-AKT1 (Phosphorylated S473) Recombinant Antibody (V2-505430) (PTM-CBMAB-0067LY)

-
Mouse Anti-FN1 Monoclonal Antibody (71) (CBMAB-1241CQ)

-
Mouse Anti-ANXA7 Recombinant Antibody (A-1) (CBMAB-A2941-YC)

-
Mouse Anti-AK4 Recombinant Antibody (V2-180419) (CBMAB-A1891-YC)

-
Mouse Anti-EMP3 Recombinant Antibody (CBFYE-0100) (CBMAB-E0207-FY)

-
Rabbit Anti-ENO2 Recombinant Antibody (BA0013) (CBMAB-0272CQ)

-
Mouse Anti-CAPZB Recombinant Antibody (CBYY-C0944) (CBMAB-C2381-YY)

-
Mouse Anti-CECR2 Recombinant Antibody (CBWJC-2465) (CBMAB-C3533WJ)

-
Mouse Anti-14-3-3 Pan Recombinant Antibody (V2-9272) (CBMAB-1181-LY)

-
Mouse Anti-BrdU Recombinant Antibody (IIB5) (CBMAB-1038CQ)

-
Rabbit Anti-CCN1 Recombinant Antibody (CBWJC-3580) (CBMAB-C4816WJ)

-
Mouse Anti-DLC1 Recombinant Antibody (D1009) (CBMAB-D1009-YC)

-
Mouse Anti-AOC3 Recombinant Antibody (CBYY-0014) (CBMAB-0014-YY)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






