RING1 Antibodies
Background
RING1 is the core component of Polycomb inhibitory complex 1 (PRC1) and is mainly involved in the epigenetic regulation of eukaryotic cells. As an E3 ubiquitin ligase, RING1 catalyzes the monoubiquitination of lysine at position 119 of histone H2A (H2AK119ub), a modification that is crucial for maintaining gene silencing during development and cell differentiation. By promoting chromatin compression, RING1 helps establish and stabilize transcriptional inhibition states, thereby precisely regulating gene expression programs. RING1 was first discovered in the 1990s when studying Polycomb family proteins in fruit flies. Subsequent studies have confirmed its conserved epigenetic silence function in mammals. Through X-ray crystallography and biochemical experiments, scientists have resolved the structure and mechanism of action of RING1, revealing how its RING domain mediates ubiquitin transfer. The dysregulation of RING1 is closely related to cancer and developmental abnormalities, highlighting its crucial role in maintaining cellular identity.
Structure of RING1
RING1, as the core component of Polycomb inhibitory complex 1 (PRC1), has a molecular weight of approximately 39-45 kDa, and the specific value varies depending on the subtype and post-translational modification. The sequence differences of key functional domains among different species may lead to subtle differences in molecular weight.
| Species | Human | Mouse | Drosophlia | Zebrafish | Arabidopsis |
| Molecular Weight (kDa) | 39.5 | 39.2 | 38.7 | 40.1 | 42.3 |
| Primary Structural Differences | Contains RING and RAWUL domains | 95% sequence homology to human | Lacks RAWUL domain | Duplicated in genome | Plant-specific N-terminal extension |
Human RING1 protein is composed of 336 amino acids with a compact tertiary structure stabilized by the RING domain (residues 50-100) and the RAWUL domain (residues 150-200). These domains mediate protein-protein interactions and E3 ubiquitin ligase activity.
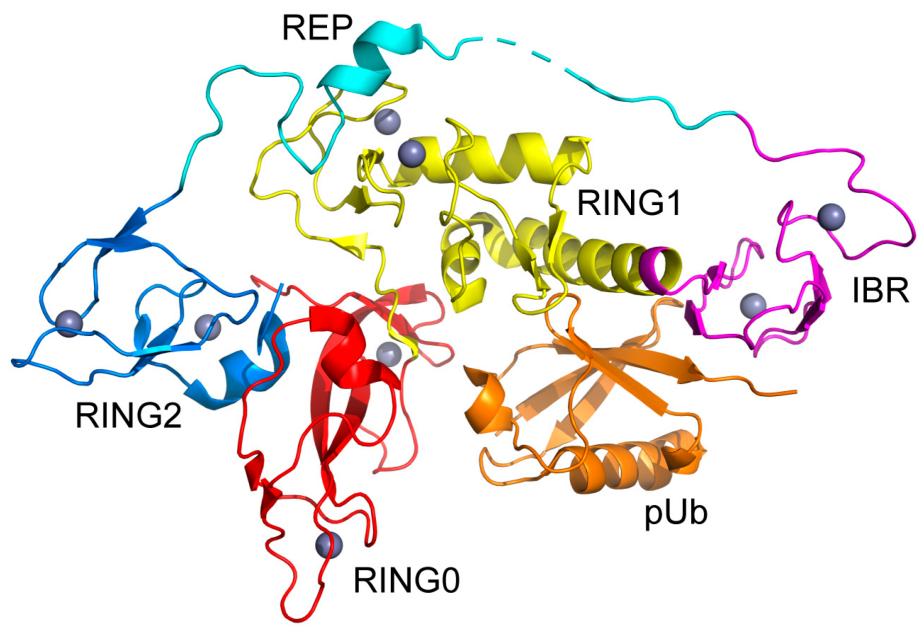 Fig. 1 Structural Insights into the RING-between-RING Architecture of Parkin Core Domain.1
Fig. 1 Structural Insights into the RING-between-RING Architecture of Parkin Core Domain.1
Key structural properties of RING1:
- Conservative RING domain
- Multi-subunit binding interface
- Dynamic control element
- Epigenetic effector module
Functions of RING1
RING1, as the core catalytic subunit of the PRC1 complex, mainly functions to regulate gene silencing through epigenetic modification. Meanwhile, it is also involved in a variety of important biological processes, including cell fate determination and the occurrence and development of diseases.
| Function | Description |
| Histone ubiquitination | As an E3 ubiquitin ligase, it catalyzes the monoubiquitination of H2AK119, directly inducing chromatin compression and transcriptional inhibition. |
| Assembly of the multicomb protein complex | Mediated by RAWUL structure domain PRC1 each sub assembly, maintain stability of complex. |
| Stem cell stemness maintenance | Silence the differentiation-related genes to ensure the self-renewal ability of pluripotent stem cells. |
| X chromosome inactivation | Synergistic with Xist RNA to establish and maintain X chromosome silence. |
| Tumorigenesis regulation | Abnormally high expression leads to the silencing of tumor suppressor genes and promotes the development of various cancers. |
The substrate recognition feature of RING1 presents a "dual-lock mechanism": Structural lock: RING-BMI1 heterodimer specifically recognizes nucleosome surfaces; Epigenetic locking: Preferentially binds to chromatin regions with pre-existing H3K27me3 markers.
Applications of RING1 and RING1 Antibody in Literature
1. Wei, Jianhua, et al. "Role of Bmi1 in H2A ubiquitylation and Hox gene silencing." Journal of Biological Chemistry 281.32 (2006): 22537-22544. https://www.jbc.org/article/S0021-9258(19)47591-1/fulltext
The article indicates that RING1 forms the H2A ubiquitin ligase complex with BMI1, RING2, etc. Among them, BMI1 significantly enhances the H2A ubiquitination activity of RING1/RING2 by stabilizing the structure of the complex. This modification directly regulates the silencing of the HoxC5 gene and cell proliferation. The deletion of BMI1 leads to the global deletion of H2A ubiquitination, the upregulation of HoxC5 expression and the slowdown of cell growth. Experiments confirmed that the ubiquitination of H2A was enriched in the 5' regulatory region of the HoxC5 gene, revealing a new mechanism by which BMI1-RING1 regulates the expression of target genes through epigenetics.
2. Gao, Song, et al. "Low expression of the polycomb protein RING1 predicts poor prognosis in human breast cancer." Frontiers in Oncology 10 (2021): 618768. https://doi.org/10.3389/fonc.2020.618768
This article's research found that RING1, a core member of the polycomb protein family, plays a key role in the occurrence and development of various cancers. As an important epigenetic regulatory factor, the expression characteristics and prognostic value of RING1 in breast cancer still need to be further explored, which provides a new research direction for targeted therapy of breast cancer.
3. Alchanati, Iris, et al. "The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2α cleavage complex–a potentially new drug target." PloS one 4.12 (2009): e8104. https://doi.org/10.1371/journal.pone.0008104
The article indicates that Bmi1/Ring1A E3 ubiquitin ligase is involved in the drug-induced degradation process of Top2α proteasome. Inhibiting Bmi1 can block the degradation of Top2α, prolong the retention time of its DNA cleavage complex, and enhance the therapeutic effect of the drug. Experiments have confirmed that this complex can directly ubiquitinate Top2α, and the small molecule inhibitors obtained through screening can block this process and produce a synergistic effect with the topoisomerase 2 agent.
4. Rui, Xiaohui, et al. "Long non-coding RNA C5orf66-AS1 promotes cell proliferation in cervical cancer by targeting miR-637/RING1 axis." Cell death & disease 9.12 (2018): 1175. https://doi.org/10.1038/s41419-018-1228-z
This article found that lncRNA C5orf66-AS1 was abnormally highly expressed in cervical cancer and relieved its inhibitory effect on RING1 by competitively binding to miR-637. RING1, as a key regulatory factor, mediates the regulatory effect of the C5orf66-AS1/miR-637 axis on the proliferation, apoptosis and cell cycle of cervical cancer cells. In vivo and in vitro experiments have confirmed that targeting this pathway can significantly inhibit tumor growth, providing a new target for the treatment of cervical cancer
5. He, Zhen, et al. "Ring 1 and YY1 binding protein is expressed in murine spermatocytes but dispensable for spermatogenesis." Genes 11.1 (2020): 84. https://doi.org/10.3390/genes11010084
This article has found that the Ring1-binding protein Rybp plays a key regulatory role in spermatogenesis. As the core component of the multicomb protein complex, RING1 is specifically enriched in spermatocytes through Rybp and regulates the expression of meiosis related genes. Although Rybp knockout does not directly affect fertility, the Ring1-RYBP complex may precisely regulate the spermatogenesis process through epigenetic mechanisms, revealing the important role of RING1 in the development of germ cells.
Creative Biolabs: RING1 Antibodies for Research
Creative Biolabs specializes in the production of high-quality RING1 antibodies for research and industrial applications. Our portfolio includes monoclonal and polyclonal antibodies tailored for ELISA, Flow Cytometry, Western Blot, Immunohistochemistry, and other research applications.
- Custom RING1 Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our RING1 antibodies, custom preparations, or technical support, contact us at info@creative-biolabs.com.
Reference
- Ye, Xiang, et al. "Cooperative Substructure and Energetics of Allosteric Regulation of the Catalytic Core of the E3 Ubiquitin Ligase Parkin by Phosphorylated Ubiquitin." Biomolecules 14.10 (2024): 1338. https://doi.org/10.3390/biom14101338
Anti-RING1 antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-CASQ1 Recombinant Antibody (CBFYC-0863) (CBMAB-C0918-FY)

-
Mouse Anti-ARHGDIA Recombinant Antibody (CBCNA-009) (CBMAB-R0415-CN)

-
Mouse Anti-DLG1 Monolconal Antibody (4F3) (CBMAB-0225-CN)

-
Mouse Anti-AKR1B1 Antibody (V2-2449) (CBMAB-1001CQ)

-
Rat Anti-AChR Recombinant Antibody (V2-12500) (CBMAB-0990-CN)

-
Human Anti-SARS-CoV-2 S1 Monoclonal Antibody (CBFYR-0120) (CBMAB-R0120-FY)

-
Mouse Anti-CAT Recombinant Antibody (724810) (CBMAB-C8431-LY)

-
Mouse Anti-CCND2 Recombinant Antibody (DCS-3) (CBMAB-G1318-LY)

-
Mouse Anti-C1QC Recombinant Antibody (CBFYC-0600) (CBMAB-C0654-FY)

-
Mouse Anti-CD8 Recombinant Antibody (C1083) (CBMAB-C1083-LY)

-
Mouse Anti-ATP1A2 Recombinant Antibody (M7-PB-E9) (CBMAB-A4013-YC)

-
Mouse Anti-AAV-5 Recombinant Antibody (V2-503417) (CBMAB-V208-1369-FY)

-
Mouse Anti-ASB9 Recombinant Antibody (1D8) (CBMAB-A0529-LY)

-
Mouse Anti-CECR2 Recombinant Antibody (CBWJC-2465) (CBMAB-C3533WJ)

-
Mouse Anti-AAV8 Recombinant Antibody (V2-634028) (CBMAB-AP022LY)

-
Mouse Anti-ADRB2 Recombinant Antibody (V2-180026) (CBMAB-A1420-YC)

-
Mouse Anti-APC Recombinant Antibody (CBYC-A661) (CBMAB-A3036-YC)

-
Mouse Anti-ACE2 Recombinant Antibody (V2-179293) (CBMAB-A0566-YC)

-
Mouse Anti-AGK Recombinant Antibody (V2-258056) (CBMAB-M0989-FY)

-
Mouse Anti-BPGM Recombinant Antibody (CBYY-1806) (CBMAB-2155-YY)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot






