SUB1 Antibodies
Background
SUB1 transcription of a gene encoding a highly conservative auxiliary activation factor protein, mainly exists in eukaryotic nuclei. This protein participates in the regulation of transcription initiation by interacting with RNA polymerase II and is involved in the coordination of DNA damage repair and cell cycle processes. Because it can enhance the activity of multiple transcription factors, SUB1 plays a key role in maintaining genomic stability and stress response. This gene was first identified in yeast in 1996, and its human homolog SUB1 (PC4) is the first mammalian protein to be confirmed to have a universal transcriptional coactivation function. Its unique DNA binding domain and the interaction mode of double-stranded/single-stranded DNA have become a classic model for studying transcriptional regulatory mechanisms, providing an important molecular basis for revealing gene expression regulation, chromatin remodeling and cancer-related signaling pathways.
Structure of SUB1
The molecular weight of the protein encoded by the SUB1 gene is approximately 14 kDa, and its precise size varies slightly among different species. This protein is composed of 127 amino acids and adopts a conserved β -folding domain as its core structure, forming a typical transcription cofactor folding pattern. Its molecular surface carries a large number of negatively charged amino acid residues, which are responsible for specific interactions with RNA polymerase II and the universal transcription factor TFIIB. The key functional region is located in the acidic activation domain at the C-terminal, which directly participates in the assembly and stabilization of the transcription initiation complex through hydrogen bonds and electrostatic interactions.
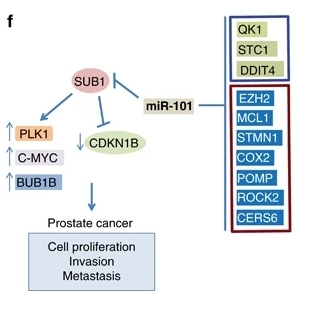 Fig. 1 Proposed model of SUB1 and miR-101 regulation in prostate cancer progression.1
Fig. 1 Proposed model of SUB1 and miR-101 regulation in prostate cancer progression.1
Key structural properties of SUB1:
- The conserved β-fold transcriptional coactivation domain was used
- The surface is rich in acidic amino acid residues
- The C-terminal acidic activation region binds RNA polymerase II and universal transcription factors
Functions of SUB1
The main function of the SUB1 gene is to act as a transcriptional coactivator to participate in the regulation of gene expression and play a key role in DNA damage repair. In addition, it also involves the regulation of cell cycle processes and stress responses.
| Function | Description |
| Transcriptional activation | SUB1 binds to RNA polymerase II and various universal transcription factors, enhancing promoter activity and promoting gene transcription initiation. |
| DNA damage repair | By participating in the nucleotide excision repair (NER) pathway, it identifies and responds to DNA damage and maintains genomic stability. |
| Cell cycle regulation | Interact with the cell cycle protein, influence the G1 / S phase transformation, regulating the process of cell proliferation. |
| Oxidative stress response | Regulate the expression of specific target genes under oxidative stress conditions to assist in maintaining intracellular REDOX balance. |
| Transcriptional fidelity is maintained | Ensure the accurate assembly of transcriptional complexes, reduce the occurrence of erroneous transcriptional events, and enhance the precision of gene expression. |
SUB1 interacts with multiple proteins through its conserved acid-activating domain to precisely regulate the basic transcriptional process in a synergistic manner. The loss of its function may lead to transcriptional abnormalities and genomic instability.
Applications of SUB1 and SUB1 Antibody in Literature
1. Huang, Rongzhong, et al. "The transcription factor SUB1 is a master regulator of the macrophage TLR response in atherosclerosis." Advanced Science 8.19 (2021): 2004162. https://doi.org/10.1002/advs.202004162
This study found that the TLR signaling pathway promotes atherosclerosis through RNA polymerase II coactivator SUB1. Myeline-specific knockout of SUB1 can enhance M2 polarization of macrophages and cholesterol effusion, and reduce plaque formation. Mechanistically, SUB1 activates Irf1 transcription in a CK2-dependent manner, driving the pro-inflammatory M1 phenotype. SUB1 is a key regulatory factor for atherosclerosis induced by TLR.
2. Liu, Gongbao, et al. "ARID1B/SUB1‐activated lncRNA HOXA‐AS2 drives the malignant behaviour of hepatoblastoma through regulation of HOXA3." Journal of cellular and molecular medicine 25.7 (2021): 3524-3536. https://doi.org/10.1111/jcmm.16435
Studies have found that the transcriptional co-activator SUB1 and the chromatin remodeling factor ARID1B jointly regulate the expression of the oncogenic lncRNA HOXA-AS2. This regulatory axis promotes the proliferation, migration and invasion of hepatoblastoma by stabilizing the HOXA3 protein. SUB1 plays a key role in the HoXa-AS2-mediated carcinogenic mechanism, providing a potential target for the treatment of hepatoblastoma.
3. Chakravarthi, Balabhadrapatruni VSK, et al. "MicroRNA-101 regulated transcriptional modulator SUB1 plays a role in prostate cancer." Oncogene 35.49 (2016): 6330-6340. https://doi.org/10.1038/onc.2016.164
Studies have found that the transcriptional co-activator SUB1 is highly expressed in aggressive prostate cancer and is negatively regulated by miR-101. SUB1 promotes its expression by binding to the promoter regions of oncogenes such as PLK1 and C-MYC, driving tumor proliferation and metastasis. Targeting SUB1 or its downstream PLK1 can inhibit tumor growth, revealing the potential of SUB1 as a new therapeutic target for prostate cancer.
4. Martinez, Mariano, et al. "Prodomain-driven enzyme dimerization: a pH-dependent autoinhibition mechanism that controls Plasmodium Sub1 activity before merozoite egress." Mbio 15.3 (2024): e00198-24. https://doi.org/10.1128/mbio.00198-24
Research has found that Plasmodium protease SUB1 regulates its spatiotemporal activation through a unique PH-dependent dimerization mechanism. Under acidic conditions, its original domain promotes the formation of inactive homodimers. It dissociates into active monomers in a neutral pH environment. This regulatory mechanism precisely controls the multi-step maturation process of SUB1 under the action of the aspartic acid protease PmX, and ultimately mediates the key step of the release of schizozoites from infected red blood cells.
5. Lidumniece, Elina, et al. "Peptidic boronic acids are potent cell-permeable inhibitors of the malaria parasite egress serine protease SUB1." Proceedings of the National Academy of Sciences 118.20 (2021): e2022696118. https://doi.org/10.1073/pnas.2022696118
Research is conducted to develop new inhibitors targeting the key protease SUB1 of the malaria parasite. This boric acid compound can effectively inhibit SUB1 activity, prevent the egress process of the parasite from red blood cells, and significantly suppress the proliferation of parasites. Such inhibitors are expected to become new antimalarial drugs with both preventive and therapeutic effects.
Creative Biolabs: SUB1 Antibodies for Research
Creative Biolabs specializes in the production of high-quality SUB1 antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom SUB1 Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our SUB1 antibodies, custom preparations, or technical support, contact us at email.
Reference
- Chakravarthi, Balabhadrapatruni VSK, et al. "MicroRNA-101 regulated transcriptional modulator SUB1 plays a role in prostate cancer." Oncogene 35.49 (2016): 6330-6340. https://doi.org/10.1038/onc.2016.164
Anti-SUB1 antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-ALB Recombinant Antibody (V2-363290) (CBMAB-S0173-CQ)

-
Mouse Anti-CRYAB Recombinant Antibody (A4345) (CBMAB-A4345-YC)

-
Mouse Anti-ADGRE5 Recombinant Antibody (V2-360335) (CBMAB-C2088-CQ)

-
Mouse Anti-Acetyl-α-Tubulin (Lys40) Recombinant Antibody (V2-623485) (CBMAB-CP2897-LY)

-
Mouse Anti-ENPP1 Recombinant Antibody (CBFYE-0159) (CBMAB-E0375-FY)

-
Rat Anti-EMCN Recombinant Antibody (28) (CBMAB-E0280-FY)

-
Mouse Anti-ASTN1 Recombinant Antibody (H-9) (CBMAB-1154-CN)

-
Rabbit Anti-ALOX5AP Recombinant Antibody (CBXF-1219) (CBMAB-F0750-CQ)

-
Mouse Anti-ATM Recombinant Antibody (2C1) (CBMAB-A3970-YC)

-
Mouse Anti-CEMIP Recombinant Antibody (3C12) (CBMAB-K0296-LY)

-
Mouse Anti-BSN Recombinant Antibody (219E1) (CBMAB-1228-CN)

-
Mouse Anti-BCL6 Recombinant Antibody (CBYY-0435) (CBMAB-0437-YY)

-
Mouse Anti-CD63 Recombinant Antibody (CBXC-1200) (CBMAB-C1467-CQ)

-
Rabbit Anti-CCN1 Recombinant Antibody (CBWJC-3580) (CBMAB-C4816WJ)

-
Mouse Anti-ALDOA Recombinant Antibody (A2) (CBMAB-A2316-YC)

-
Mouse Anti-dsDNA Recombinant Antibody (22) (CBMAB-AP1954LY)

-
Mouse Anti-BCL6 Recombinant Antibody (CBYY-0442) (CBMAB-0445-YY)

-
Mouse Anti-ADGRL2 Recombinant Antibody (V2-58519) (CBMAB-L0166-YJ)

-
Mouse Anti-EPO Recombinant Antibody (CBFYR0196) (CBMAB-R0196-FY)

-
Mouse Anti-CTNND1 Recombinant Antibody (CBFYC-2414) (CBMAB-C2487-FY)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot








